Conserved Structure and Evolution of DPF Domain of PHF10-The Specific Subunit of PBAF Chromatin Remodeling Complex
- PMID: 34681795
- PMCID: PMC8538644
- DOI: 10.3390/ijms222011134
Conserved Structure and Evolution of DPF Domain of PHF10-The Specific Subunit of PBAF Chromatin Remodeling Complex
Abstract
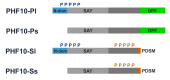
Transcription activation factors and multisubunit coactivator complexes get recruited at specific chromatin sites via protein domains that recognize histone modifications. Single PHDs (plant homeodomains) interact with differentially modified H3 histone tails. Double PHD finger (DPF) domains possess a unique structure different from PHD and are found in six proteins: histone acetyltransferases MOZ and MORF; chromatin remodeling complex BAF (DPF1-3); and chromatin remodeling complex PBAF (PHF10). Among them, PHF10 stands out due to the DPF sequence, structure, and functions. PHF10 is ubiquitously expressed in developing and adult organisms as four isoforms differing in structure (the presence or absence of DPF) and transcription regulation functions. Despite the importance of the DPF domain of PHF10 for transcription activation, its structure remains undetermined. We performed homology modeling of the human PHF10 DPF domain and determined common and distinct features in structure and histone modifications recognition capabilities, which can affect PBAF complex chromatin recruitment. We also traced the evolution of DPF1-3 and PHF10 genes from unicellular to vertebrate organisms. The data reviewed suggest that the DPF domain of PHF10 plays an important role in SWI/SNF-dependent chromatin remodeling during transcription activation.
Keywords: DPF domains; H3K14ac; PBAF; PHD; PHF10; chromatin remodeling; domain evolution; duplication; genes activation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Jain K., Fraser C.S., Marunde M.R., Parker M.M., Sagum C., Burg J.M., Hall N., Popova I.K., Rodriguez K.L., Vaidya A., et al. Characterization of the Plant Homeodomain (PHD) Reader Family for Their Histone Tail Interactions. Epigenetics Chromatin. 2020;13:3. doi: 10.1186/s13072-020-0328-z. - DOI - PMC - PubMed
-
- Morrison E.A., Musselman C.A. Chromatin Signaling and Diseases. Academic Press; London, UK: 2016. The Role of PHD Fingers in Chromatin Signaling: Mechanisms and Functional Consequences of the Recognition of Histone and Non-histone Targets; pp. 127–147.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

