Polygenic Risk Scores Contribute to Personalized Medicine of Parkinson's Disease
- PMID: 34683174
- PMCID: PMC8539098
- DOI: 10.3390/jpm11101030
Polygenic Risk Scores Contribute to Personalized Medicine of Parkinson's Disease
Abstract
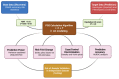
Parkinson's disease (PD) is the second most common neurodegenerative disorder characterized by the loss of dopaminergic neurons. The vast majority of PD patients develop the disease sporadically and it is assumed that the cause lies in polygenic and environmental components. The overall polygenic risk is the result of a large number of common low-risk variants discovered by large genome-wide association studies (GWAS). Polygenic risk scores (PRS), generated by compiling genome-wide significant variants, are a useful prognostic tool that quantifies the cumulative effect of genetic risk in a patient and in this way helps to identify high-risk patients. Although there are limitations to the construction and application of PRS, such as considerations of limited genetic underpinning of diseases explained by SNPs and generalizability of PRS to other populations, this personalized risk prediction could make a promising contribution to stratified medicine and tailored therapeutic interventions in the future.
Keywords: Parkinson’s disease; personalized medicine; polygenic risk scores.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Nalls M.A., Blauwendraat C., Vallerga C.L., Heilbron K., Bandres-Ciga S., Chang D., Tan M., Kia D.A., Noyce A.J., Xue A. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

