A Multi-Point Surveillance for Antimicrobial Resistance Profiles among Clinical Isolates of Gram-Negative Bacteria Recovered from Major Ha'il Hospitals, Saudi Arabia
- PMID: 34683344
- PMCID: PMC8537776
- DOI: 10.3390/microorganisms9102024
A Multi-Point Surveillance for Antimicrobial Resistance Profiles among Clinical Isolates of Gram-Negative Bacteria Recovered from Major Ha'il Hospitals, Saudi Arabia
Abstract
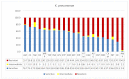
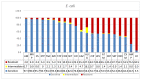
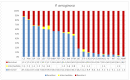
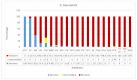
The devastating nosocomial resistance is an on-going global concern. Surveillance of resistance is crucial for efficient patient care. This study was aimed to conduct a surveillance in four major Ha'il Hospitals from September to December 2020. Using a multipoint program, records of 621 non-duplicate Gram-negative cultures were tested across 21 drugs belonging to different categories. Major species were Klebsiella pneumoniae (n = 187, 30%), E. coli (n = 151, 24.5%), Pseudomonas aeruginosa, (n = 84, 13.6%), Acinetobacter baumannii (n = 82, 13.3%), and Proteus mirabilis (n = 46, 7%). Based on recent resistance classifications, A. baumanni, P. aeruginosa, and enteric bacteria were defined as pan-resistant, extremely resistant, and multi-drug resistant, respectively. A. baumannii (35%) and K. pneumoniae (23%) dominated among coinfections in SARS-CoV2 patients. The "other Gram-negative bacteria" (n = 77, 12.5%) from diverse sources showed unique species-specific resistance patterns, while sharing a common Gram-negative resistance profile. Among these, Providencia stuartii was reported for the first time in Ha'il. In addition, specimen source, age, and gender differences played significant roles in susceptibility. Overall infection rates were 30% in ICU, 17.5% in medical wards, and 13.5% in COVID-19 zones, mostly in male (59%) senior (54%) patients. In ICU, infections were caused by P. mirabilis (52%), A. baumannii (49%), P. aeruginosa (41%), K. pneumoniae (24%), and E. coli (21%), and most of the respiratory infections were caused by carbapenem-resistant A. baumannii and K. pneumoniae and UTI by K. pneumoniae and E. coli. While impressive IC, hospital performances, and alternative treatment options still exist, the spread of resistant Gram-negative bacteria is concerning especially in geriatric patients. The high selective SARS-CoV2 coinfection by A. baumannii and K. pneumoniae, unlike the low global rates, warrants further vertical studies. Attributes of resistances are multifactorial in Saudi Arabia because of its global partnership as the largest economic and pilgrimage hub with close social and cultural ties in the region, especially during conflicts and political unrests. However, introduction of advanced inter-laboratory networks for genome-based surveillances is expected to reduce nosocomial resistances.
Keywords: Gram-negative bacterial resistances; antimicrobial-resistance surveillance; multifactorial nosocomial resistances; nosocomial-resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Mshana S.E., Gerwing L., Minde M., Hain T., Domann E., Lyamuya E., Chakraborty T., Imirzalioglu C. Outbreak of a novel Enterobacter sp. carrying bla CTX-M-15 in a neonatal unit of a tertiary care hospital in Tanzania. Int. J. Antimicrob. Agents. 2011;38:265–269. - PubMed
-
- WHO Antimicrobial Resistance: Global Report on Surveillance. [(accessed on 17 October 2019)]. Available online: https://scholar.google.de/scholar?hl=en&q=Antimicrobial+resistance%3A+gl...
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

