Lipid Profiling of Alzheimer's Disease Brain Highlights Enrichment in Glycerol(phospho)lipid, and Sphingolipid Metabolism
- PMID: 34685570
- PMCID: PMC8534054
- DOI: 10.3390/cells10102591
Lipid Profiling of Alzheimer's Disease Brain Highlights Enrichment in Glycerol(phospho)lipid, and Sphingolipid Metabolism
Abstract
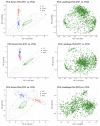
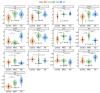
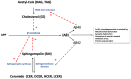
Alzheimer's disease (AD) is reported to be closely linked with abnormal lipid metabolism. To gain a more comprehensive understanding of what causes AD and its subsequent development, we profiled the lipidome of postmortem (PM) human brains (neocortex) of people with a range of AD pathology (Braak 0-6). Using high-resolution mass spectrometry, we employed a semi-targeted, fully quantitative lipidomics profiling method (Lipidyzer) to compare the biochemical profiles of brain tissues from persons with mild AD (n = 15) and severe AD (AD; n = 16), and compared them with age-matched, cognitively normal controls (n = 16). Univariate analysis revealed that the concentrations of 420 lipid metabolites significantly (p < 0.05; q < 0.05) differed between AD and controls. A total of 49 lipid metabolites differed between mild AD and controls, and 439 differed between severe AD and mild AD. Interestingly, 13 different subclasses of lipids were significantly perturbed, including neutral lipids, glycerolipids, glycerophospholipids, and sphingolipids. Diacylglycerol (DAG) (14:0/14:0), triacylglycerol (TAG) (58:10/FA20:5), and TAG (48:4/FA18:3) were the most notably altered lipids when AD and control brains were compared (p < 0.05). When we compare mild AD and control brains, phosphatidylethanolamine (PE) (p-18:0/18:1), phosphatidylserine (PS) (18:1/18:2), and PS (14:0/22:6) differed the most (p < 0.05). PE (p-18:0/18:1), DAG (14:0/14:0), and PS (18:1/20:4) were identified as the most significantly perturbed lipids when AD and mild AD brains were compared (p < 0.05). Our analysis provides the most extensive lipid profiling yet undertaken in AD brain tissue and reveals the cumulative perturbation of several lipid pathways with progressive disease pathology. Lipidomics has considerable potential for studying AD etiology and identifying early diagnostic biomarkers.
Keywords: Alzheimer’s disease; brain; lipidomics; metabolomics; pathogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–944. doi: 10.1212/WNL.34.7.939. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

