SP-1, a Serine Protease from the Gut Microbiota, Influences Colitis and Drives Intestinal Dysbiosis in Mice
- PMID: 34685638
- PMCID: PMC8534766
- DOI: 10.3390/cells10102658
SP-1, a Serine Protease from the Gut Microbiota, Influences Colitis and Drives Intestinal Dysbiosis in Mice
Abstract
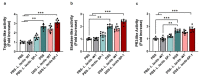
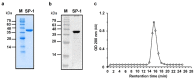
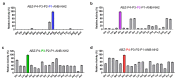
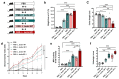
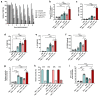
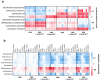
Increased protease activity has been linked to the pathogenesis of IBD. While most studies have been focusing on host proteases in gut inflammation, it remains unclear how to address the potential contribution of their bacterial counterparts. In the present study, we report a functional characterization of a newly identified serine protease, SP-1, from the human gut microbiota. The serine protease repertoire of gut Clostridium was first explored, and the specificity of SP-1 was analyzed using a combinatorial chemistry method. Combining in vitro analyses and a mouse model of colitis, we show that oral administration of recombinant bacteria secreting SP-1 (i) compromises the epithelial barrier, (ii) alters the microbial community, and (ii) exacerbates colitis. These findings suggest that gut microbial protease activity may constitute a valuable contributor to IBD and could, therefore, represent a promising target for the treatment of the disease.
Keywords: gut microbiota; holobiont; inflammatory bowel diseases; microbiome; serine proteases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

