Environmental and Genetic Factors Affecting Apospory Expressivity in Diploid Paspalum rufum
- PMID: 34685909
- PMCID: PMC8537111
- DOI: 10.3390/plants10102100
Environmental and Genetic Factors Affecting Apospory Expressivity in Diploid Paspalum rufum
Abstract
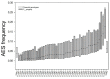
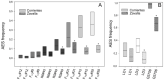
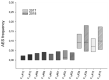
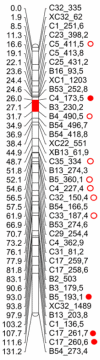
In angiosperms, gametophytic apomixis (clonal reproduction through seeds) is strongly associated with polyploidy and hybridization. The trait is facultative and its expressivity is highly variable between genotypes. Here, we used an F1 progeny derived from diploid apomictic (aposporic) genotypes of Paspalum rufum and two F2 families, derived from F1 hybrids with different apospory expressivity (%AES), to analyze the influence of the environment and the transgenerational transmission of the trait. In addition, AFLP markers were developed in the F1 population to identify genomic regions associated with the %AES. Cytoembryological analyses showed that the %AES was significantly influenced by different environments, but remained stable across the years. F1 and F2 progenies showed a wide range of %AES variation, but most hybrids were not significantly different from the parental genotypes. Maternal and paternal genetic linkage maps were built covering the ten expected linkage groups (LG). A single-marker analysis detected at least one region of 5.7 cM on LG3 that was significantly associated with apospory expressivity. Our results underline the importance of environmental influence in modulating apospory expressivity and identified a genomic region associated with apospory expressivity at the diploid level.
Keywords: Paspalum rufum; apospory expressivity; diploid level; environment; inheritance.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Kumar S. Epigenetic control of apomixis: A new perspective of an old enigma. Adv. Plants Agric. Res. 2017;7:10-15406. doi: 10.15406/apar.2017.07.00243. - DOI
-
- Toenniessen G.H. Feeding the world in the 21st century: Plant breeding, biotechnology, and the potential role of apomixis. In: Savidan Y., Carman J.G., Dresselhaus T., editors. The Flowering of Apomixis: From Mechanisms to Genetic Engineering. CIMMYT, IRD, European Commission DG VI (FAIR); Mexico City, Mexico: 2001. pp. 1–7.
-
- Whitton J., Sears C.J., Baack E.J., Otto S.P. The dynamic nature of apomixis in the angiosperms. Int. J. Plant Sci. 2008;169:169–182. doi: 10.1086/523369. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

