Comprehensive Analyses of NAC Transcription Factor Family in Almond (Prunus dulcis) and Their Differential Gene Expression during Fruit Development
- PMID: 34686009
- PMCID: PMC8541688
- DOI: 10.3390/plants10102200
Comprehensive Analyses of NAC Transcription Factor Family in Almond (Prunus dulcis) and Their Differential Gene Expression during Fruit Development
Abstract
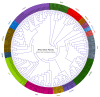
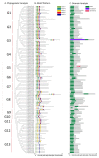
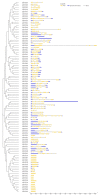
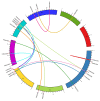
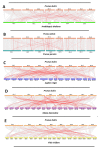
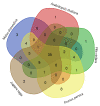
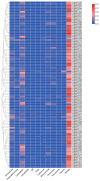
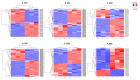
As plant specific transcription factors, NAC (NAM, ATAF1/2, CUC2) domain is involved in the plant development and stress responses. Due to the vitality of NAC gene family, BLASTp was performed to identify NAC genes in almond (Prunus dulcis). Further, phylogenetic and syntenic analyses were performed to determine the homology and evolutionary relationship. Gene duplication, gene structure, motif, subcellular localization, and cis-regulatory analyses were performed to assess the function of PdNAC. Whereas RNA-seq analysis was performed to determine the differential expression of PdNAC in fruits at various developmental stages. We identified 106 NAC genes in P. dulcis genome and were renamed according to their chromosomal distribution. Phylogenetic analysis in both P. dulcis and Arabidopsis thaliana revealed the presence of 14 subfamilies. Motif and gene structure followed a pattern according to the PdNAC position in phylogenetic subfamilies. Majority of NAC are localized in the nucleus and have ABA-responsive elements in the upstream region of PdNAC. Differential gene expression analyses revealed one and six PdNAC that were up and down-regulated, respectively, at all development stages. This study provides insights into the structure and function of PdNAC along with their role in the fruit development to enhance an understanding of NAC in P. dulcis.
Keywords: NAC; RNA-seq; differential gene expression; gene duplication; genome wide identification; phylogenetic analysis; syntenic analysis; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials

