Targeting miR-126 in inv(16) acute myeloid leukemia inhibits leukemia development and leukemia stem cell maintenance
- PMID: 34686664
- PMCID: PMC8536759
- DOI: 10.1038/s41467-021-26420-7
Targeting miR-126 in inv(16) acute myeloid leukemia inhibits leukemia development and leukemia stem cell maintenance
Abstract
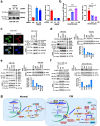
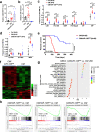
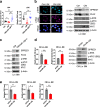
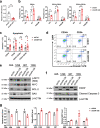
Acute myeloid leukemia (AML) harboring inv(16)(p13q22) expresses high levels of miR-126. Here we show that the CBFB-MYH11 (CM) fusion gene upregulates miR-126 expression through aberrant miR-126 transcription and perturbed miR-126 biogenesis via the HDAC8/RAN-XPO5-RCC1 axis. Aberrant miR-126 upregulation promotes survival of leukemia-initiating progenitors and is critical for initiating and maintaining CM-driven AML. We show that miR-126 enhances MYC activity through the SPRED1/PLK2-ERK-MYC axis. Notably, genetic deletion of miR-126 significantly reduces AML rate and extends survival in CM knock-in mice. Therapeutic depletion of miR-126 with an anti-miR-126 (miRisten) inhibits AML cell survival, reduces leukemia burden and leukemia stem cell (LSC) activity in inv(16) AML murine and xenograft models. The combination of miRisten with chemotherapy further enhances the anti-leukemia and anti-LSC activity. Overall, this study provides molecular insights for the mechanism and impact of miR-126 dysregulation in leukemogenesis and highlights the potential of miR-126 depletion as a therapeutic approach for inv(16) AML.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

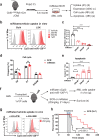
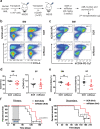
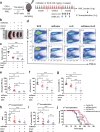
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

