A human multi-lineage hepatic organoid model for liver fibrosis
- PMID: 34686668
- PMCID: PMC8536785
- DOI: 10.1038/s41467-021-26410-9
A human multi-lineage hepatic organoid model for liver fibrosis
Abstract
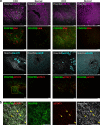
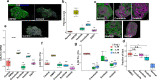
To investigate the pathogenesis of a congenital form of hepatic fibrosis, human hepatic organoids were engineered to express the most common causative mutation for Autosomal Recessive Polycystic Kidney Disease (ARPKD). Here we show that these hepatic organoids develop the key features of ARPKD liver pathology (abnormal bile ducts and fibrosis) in only 21 days. The ARPKD mutation increases collagen abundance and thick collagen fiber production in hepatic organoids, which mirrors ARPKD liver tissue pathology. Transcriptomic and other analyses indicate that the ARPKD mutation generates cholangiocytes with increased TGFβ pathway activation, which are actively involved stimulating myofibroblasts to form collagen fibers. There is also an expansion of collagen-producing myofibroblasts with markedly increased PDGFRB protein expression and an activated STAT3 signaling pathway. Moreover, the transcriptome of ARPKD organoid myofibroblasts resemble those present in commonly occurring forms of liver fibrosis. PDGFRB pathway involvement was confirmed by the anti-fibrotic effect observed when ARPKD organoids were treated with PDGFRB inhibitors. Besides providing insight into the pathogenesis of congenital (and possibly acquired) forms of liver fibrosis, ARPKD organoids could also be used to test the anti-fibrotic efficacy of potential anti-fibrotic therapies.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

