Dichotomous metabolic networks govern human ILC2 proliferation and function
- PMID: 34686862
- PMCID: PMC8553616
- DOI: 10.1038/s41590-021-01043-8
Dichotomous metabolic networks govern human ILC2 proliferation and function
Abstract
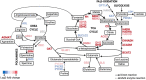
Group 2 innate lymphoid cells (ILC2s) represent innate homologs of type 2 helper T cells (TH2) that participate in immune defense and tissue homeostasis through production of type 2 cytokines. While T lymphocytes metabolically adapt to microenvironmental changes, knowledge of human ILC2 metabolism is limited, and its key regulators are unknown. Here, we show that circulating 'naive' ILC2s have an unexpected metabolic profile with a higher level of oxidative phosphorylation (OXPHOS) than natural killer (NK) cells. Accordingly, ILC2s are severely reduced in individuals with mitochondrial disease (MD) and impaired OXPHOS. Metabolomic and nutrient receptor analysis revealed ILC2 uptake of amino acids to sustain OXPHOS at steady state. Following activation with interleukin-33 (IL-33), ILC2s became highly proliferative, relying on glycolysis and mammalian target of rapamycin (mTOR) to produce IL-13 while continuing to fuel OXPHOS with amino acids to maintain cellular fitness and proliferation. Our results suggest that proliferation and function are metabolically uncoupled in human ILC2s, offering new strategies to target ILC2s in disease settings.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











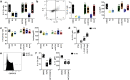
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

