Glioma: molecular signature and crossroads with tumor microenvironment
- PMID: 34687436
- PMCID: PMC8924130
- DOI: 10.1007/s10555-021-09997-9
Glioma: molecular signature and crossroads with tumor microenvironment
Abstract
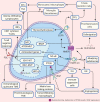
In patients with glioblastoma, the average survival time with current treatments is short, mainly due to recurrences and resistance to therapy. This insufficient treatment success is, in large parts, due to the tremendous molecular heterogeneity of gliomas, which affects the overall prognosis and response to therapies and plays a vital role in gliomas' grading. In addition, the tumor microenvironment is a major player for glioma development and resistance to therapy. Active communication between glioma cells and local or neighboring healthy cells and the immune environment promotes the cancerogenic processes and contributes to establishing glioma stem cells, which drives therapy resistance. Besides genetic alterations in the primary tumor, tumor-released factors, cytokines, proteins, extracellular vesicles, and environmental influences like hypoxia provide tumor cells the ability to evade host tumor surveillance machinery and promote disease progression. Moreover, there is increasing evidence that these players affect the molecular biological properties of gliomas and enable inter-cell communication that supports pro-cancerogenic cell properties. Identifying and characterizing these complex mechanisms are inevitably necessary to adapt therapeutic strategies and to develop novel measures. Here we provide an update about these junctions where constant traffic of biomolecules adds complexity in the management of glioblastoma.
Keywords: Cancer microenvironment; Cancer stem cells; Glioblastoma; Glioma; Stem cells; Tumor microenvironment.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

