Prolonged Impairment of Short-Chain Fatty Acid and L-Isoleucine Biosynthesis in Gut Microbiome in Patients With COVID-19
- PMID: 34687739
- PMCID: PMC8529231
- DOI: 10.1053/j.gastro.2021.10.013
Prolonged Impairment of Short-Chain Fatty Acid and L-Isoleucine Biosynthesis in Gut Microbiome in Patients With COVID-19
Abstract
Background and aims: Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is associated with altered gut microbiota composition. Phylogenetic groups of gut bacteria involved in the metabolism of short chain fatty acids (SCFAs) were depleted in SARS-CoV-2-infected patients. We aimed to characterize a functional profile of the gut microbiome in patients with COVID-19 before and after disease resolution.
Methods: We performed shotgun metagenomic sequencing on fecal samples from 66 antibiotics-naïve patients with COVID-19 and 70 non-COVID-19 controls. Serial fecal samples were collected (at up to 6 times points) during hospitalization and beyond 1 month after discharge. We assessed gut microbial pathways in association with disease severity and blood inflammatory markers. We also determined changes of microbial functions in fecal samples before and after disease resolution and validated these functions using targeted analysis of fecal metabolites.
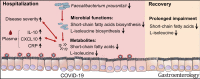
Results: Compared with non-COVID-19 controls, patients with COVID-19 with severe/critical illness showed significant alterations in gut microbiome functionality (P < .001), characterized by impaired capacity of gut microbiome for SCFA and L-isoleucine biosynthesis and enhanced capacity for urea production. Impaired SCFA and L-isoleucine biosynthesis in gut microbiome persisted beyond 30 days after recovery in patients with COVID-19. Targeted analysis of fecal metabolites showed significantly lower fecal concentrations of SCFAs and L-isoleucine in patients with COVID-19 before and after disease resolution. Lack of SCFA and L-isoleucine biosynthesis significantly correlated with disease severity and increased plasma concentrations of CXCL-10, NT- proB-type natriuretic peptide, and C-reactive protein (all P < .05).
Conclusions: Gut microbiome of patients with COVID-19 displayed impaired capacity for SCFA and L-isoleucine biosynthesis that persisted even after disease resolution. These 2 microbial functions correlated with host immune response underscoring the importance of gut microbial functions in SARS-CoV-2 infection pathogenesis and outcome.
Keywords: Coronavirus; Gut Microbiome; Microbial Functions; SCFAs.
Copyright © 2022 AGA Institute. Published by Elsevier Inc. All rights reserved.
Figures









Comment in
-
COVID-19 and the Forgotten Organ: Prolonged Changes to the Metabolic Output of the Gut Microbiome.Gastroenterology. 2022 Feb;162(2):394-396. doi: 10.1053/j.gastro.2021.11.017. Epub 2021 Nov 17. Gastroenterology. 2022. PMID: 34800482 Free PMC article. No abstract available.
-
Gut Microbiome Metabolism Drives the Resolution of Patients With Coronavirus Disease 2019.Gastroenterology. 2022 Jul;163(1):336-337. doi: 10.1053/j.gastro.2021.12.261. Epub 2021 Dec 23. Gastroenterology. 2022. PMID: 34954225 Free PMC article. No abstract available.
References
-
- Yu L., Tong Y., Shen G., et al. Immunodepletion with hypoxemia: a potential high risk subtype of coronavirus disease 2019. MedRxiv. 2020 03.03.20030650.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

