Translational suppression of SARS-COV-2 ORF8 protein mRNA as a Viable therapeutic target against COVID-19: Computational studies on potential roles of isolated compounds from Clerodendrum volubile leaves
- PMID: 34688170
- PMCID: PMC8524706
- DOI: 10.1016/j.compbiomed.2021.104964
Translational suppression of SARS-COV-2 ORF8 protein mRNA as a Viable therapeutic target against COVID-19: Computational studies on potential roles of isolated compounds from Clerodendrum volubile leaves
Abstract
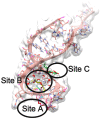
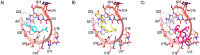
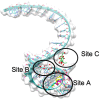
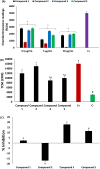
The open reading frame 8 (ORF8) protein of SARS-CoV-2 has been implicated in the onset of cytokine storms, which are responsible for the pathophysiology of COVID-19 infection. The present study investigated the potential of isolated compounds from Clerodendrum volubile leaves to stall oxidative bursts in vitro and interact with ORF8 mRNA segments of the SARS-CoV-2 whole genome using computational tools. Five compounds, namely, harpagide, 1-(3-methyl-2-butenoxy)-4-(1-propenyl)benzene, ajugoside, iridoid glycoside and erucic acid, were isolated from C. volubile leaves, and their structures were elucidated using conventional spectroscopy tools. Iridoid glycoside is being reported for the first time and is thus regarded as a new compound. The ORF8 mRNA sequences of the translation initiation sites (TIS) and translation termination sites (TTSs) encoding ORF8 amino acids were retrieved from the full genome of SARS-CoV-2. Molecular docking studies revealed strong molecular interactions of the isolated compounds with the TIS and TTS of ORF8 mRNA. Harpagide showed the strongest binding affinity for TIS, while erucic acid was the strongest for TTS. The immunomodulatory potentials of the isolated compounds were investigated on neutrophil phagocytic respiratory bursts using luminol-amplified chemiluminescence technique. The compounds significantly inhibited oxidative burst, with 1-(3-methyl-2-butenoxy)-4-(1-propenyl)benzene having the best activity. Ajugoside and erucic acid showed significant inhibitory activity on T-cell proliferation. These results indicate the potential of C. volubile compounds as immunomodulators and can be utilized to curb cytokine storms implicated in COVID-19 infection. These potentials are further corroborated by the strong interactions of the compounds with the TIS and TTS of ORF8 mRNA from the SARS-CoV-2 whole genome.
Keywords: And SARS-CoV-2; COVID-19; Clerodendrum volubile; Cytokine storm; Iridoid glycoside; ORF8.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
None Declared.
Figures







References
-
- Parvez M.K., Jagirdar R.M., Purty R.S., Venkata S.K., Agrawal V., Kumar J., Tiwari N. COVID-19 pandemic: understanding the emergence, pathogenesis and containment. World Acad. Sci. J. 2020;2(5) 1-1.
-
- Phelan A.L., Katz R., Gostin L.O. The novel coronavirus originating in Wuhan, China: challenges for global health governance. Jama. 2020;323(8):709–710. - PubMed
-
- James N., Menzies M., Radchenko P. COVID-19 second wave mortality in Europe and the United States. Chaos. 2021;31(3) - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

