TAGOPSIN: collating taxa-specific gene and protein functional and structural information
- PMID: 34688246
- PMCID: PMC8541804
- DOI: 10.1186/s12859-021-04429-5
TAGOPSIN: collating taxa-specific gene and protein functional and structural information
Abstract
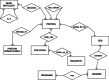
Background: The wealth of biological information available nowadays in public databases has triggered an unprecedented rise in multi-database search and data retrieval for obtaining detailed information about key functional and structural entities. This concerns investigations ranging from gene or genome analysis to protein structural analysis. However, the retrieval of interconnected data from a number of different databases is very often done repeatedly in an unsystematic way.
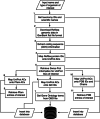
Results: Here, we present TAxonomy, Gene, Ontology, Protein, Structure INtegrated (TAGOPSIN), a command line program written in Java for rapid and systematic retrieval of select data from seven of the most popular public biological databases relevant to comparative genomics and protein structure studies. The program allows a user to retrieve organism-centred data and assemble them in a single data warehouse which constitutes a useful resource for several biological applications. TAGOPSIN was tested with a number of organisms encompassing eukaryotes, prokaryotes and viruses. For example, it successfully integrated data for about 17,000 UniProt entries of Homo sapiens and 21 UniProt entries of human coronavirus.
Conclusion: TAGOPSIN demonstrates efficient data integration whereby manipulation of interconnected data is more convenient than doing multi-database queries. The program facilitates for instance interspecific comparative analyses of protein-coding genes in a molecular evolutionary study, or identification of taxa-specific protein domains and three-dimensional structures. TAGOPSIN is available as a JAR file at https://github.com/ebundhoo/TAGOPSIN and is released under the GNU General Public License.
Keywords: Comparative genomics; Data integration; Data retrieval; Database; Object-oriented biology.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Burley SK, Berman HM, Bhikadiya C, Bi C, Chen L, Di Costanzo L, Christie C, Dalenberg K, Duarte JM, Dutta S, Feng Z, Ghosh S, Goodsell DS, Green RK, Guranovic V, Guzenko D, Hudson BP, Kalro T, Liang Y, Lowe R, Namkoong H, Peisach E, Periskova I, Prlic A, Randle C, Rose A, Rose P, Sala R, Sekharan M, Shao C, Tan L, Tao YP, Valasatava Y, Voigt M, Westbrook J, Woo J, Yang H, Young J, Zhuravleva M, Zardecki C. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47(D1):464–74. doi: 10.1093/nar/gky1004. - DOI - PMC - PubMed
-
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):427–32. doi: 10.1093/nar/gky995. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

