Comprehensive characterization of internal and cuticle surface microbiota of laboratory-reared F1 Anopheles albimanus originating from different sites
- PMID: 34688298
- PMCID: PMC8542342
- DOI: 10.1186/s12936-021-03934-5
Comprehensive characterization of internal and cuticle surface microbiota of laboratory-reared F1 Anopheles albimanus originating from different sites
Abstract
Background: Research on mosquito-microbe interactions may lead to new tools for mosquito and mosquito-borne disease control. To date, such research has largely utilized laboratory-reared mosquitoes that typically lack the microbial diversity of wild populations. A logical progression in this area involves working under controlled settings using field-collected mosquitoes or, in most cases, their progeny. Thus, an understanding of how laboratory colonization affects the assemblage of mosquito microbiota would aid in advancing mosquito microbiome studies and their applications beyond laboratory settings.
Methods: Using high throughput 16S rRNA amplicon sequencing, the internal and cuticle surface microbiota of F1 progeny of wild-caught adult Anopheles albimanus from four locations in Guatemala were characterized. A total of 132 late instar larvae and 135 2-5 day-old, non-blood-fed virgin adult females that were reared under identical laboratory conditions, were pooled (3 individuals/pool) and analysed.
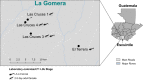
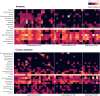
Results: Results showed location-associated heterogeneity in both F1 larval internal (p = 0.001; pseudo-F = 9.53) and cuticle surface (p = 0.001; pseudo-F = 8.51) microbiota, and only F1 adult cuticle surface (p = 0.001; pseudo-F = 4.5) microbiota, with a more homogenous adult internal microbiota (p = 0.12; pseudo-F = 1.6) across collection sites. Overall, ASVs assigned to Leucobacter, Thorsellia, Chryseobacterium and uncharacterized Enterobacteriaceae, dominated F1 larval internal microbiota, while Acidovorax, Paucibacter, and uncharacterized Comamonadaceae, dominated the larval cuticle surface. F1 adults comprised a less diverse microbiota compared to larvae, with ASVs assigned to the genus Asaia dominating both internal and cuticle surface microbiota, and constituting at least 70% of taxa in each microbial niche.
Conclusions: These results suggest that location-specific heterogeneity in filed mosquito microbiota can be transferred to F1 progeny under normal laboratory conditions, but this may not last beyond the F1 larval stage without adjustments to maintain field-derived microbiota. These findings provide the first comprehensive characterization of laboratory-colonized F1 An. albimanus progeny from field-derived mothers. This provides a background for studying how parentage and environmental conditions differentially or concomitantly affect mosquito microbiome composition, and how this can be exploited in advancing mosquito microbiome studies and their applications beyond laboratory settings.
Keywords: 16S rRNA gene amplicon sequencing; Anopheles albimanus; Laboratory colonization; Mosquito microbiome; Mosquito microbiota; Next generation sequencing.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Lindh JM, Borg-Karlson AK, Faye I. Transstadial and horizontal transfer of bacteria within a colony of Anopheles gambiae (Diptera: Culicidae) and oviposition response to bacteria-containing water. Acta Trop. 2008;107:242–250. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

