The intrinsic kinase activity of BRD4 spans its BD2-B-BID domains
- PMID: 34688663
- PMCID: PMC8591364
- DOI: 10.1016/j.jbc.2021.101326
The intrinsic kinase activity of BRD4 spans its BD2-B-BID domains
Abstract
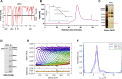
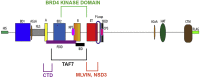
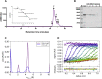
Bromodomain protein 4 (BRD4) is a transcriptional and epigenetic regulator that is a therapeutic target in many cancers and inflammatory diseases. BRD4 plays important roles in transcription as an active kinase, which phosphorylates the carboxy-terminal domain (CTD) of RNA polymerase II (Pol II), the proto-oncogene c-MYC, and transcription factors TAF7 and CDK9. BRD4 is also a passive scaffold that recruits transcription factors. Despite these well-established functions, there has been little characterization of BRD4's biophysical properties or its kinase activity. We report here that the 156 kD mouse BRD4 exists in an extended dimeric conformation with a sedimentation coefficient of ∼6.7 S and a high frictional ratio. Deletion of the conserved B motif (aa 503-548) disrupts BRD4's dimerization. BRD4 kinase activity maps to amino acids 351 to 598, which span bromodomain-2, the B motif, and the BID domain (BD2-B-BID) and contributes to the in vivo phosphorylation of its substrates. As further assessed by analytical ultracentrifugation, BRD4 directly binds purified Pol II CTD. Importantly, the conserved A motif of BRD4 is essential for phosphorylation of Pol II CTD, but not for phosphorylation of TAF7, mapping its binding site to the A motif. Peptides of the viral MLV integrase (MLVIN) protein and cellular histone lysine methyltransferase, NSD3, which have been shown by NMR to bind to the extra-terminal (ET) domain, also are phosphorylated by BRD4. Thus, BRD4 has multiple distinct substrate-binding sites and a common kinase domain. These results provide new insights into the structure and kinase function of BRD4.
Keywords: BRD4; CTD; extended dimer; kinase.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

