ARID1A deficiency weakens BRG1-RAD21 interaction that jeopardizes chromatin compactness and drives liver cancer cell metastasis
- PMID: 34689165
- PMCID: PMC8542038
- DOI: 10.1038/s41419-021-04291-6
ARID1A deficiency weakens BRG1-RAD21 interaction that jeopardizes chromatin compactness and drives liver cancer cell metastasis
Abstract
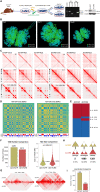
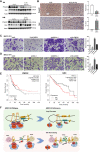
ARID1A, encoding a subunit of SWI/SNF chromatin remodeling complex, is widely recognized as a tumor suppressor gene in multiple tumor types including liver cancer. Previous studies have demonstrated that ARID1A deficiency can cause liver cancer metastasis, possibly due to the altered chromatin organization, however the underlying mechanisms remain poorly understood. To address the effect of Arid1a deficiency on chromatin organization, we generated chromatin interaction matrices, and exploited the conformation changes upon Arid1a depletion in hepatocytes. Our results demonstrated that Arid1a deficiency induced A/B compartment switching, topologically associated domain (TAD) remodeling, and decrease of chromatin loops. Further mechanism studies revealed that ATPase BRG1 of SWI/SNF complex could physically interact with RAD21, a structural subunit of chromatin architectural element cohesin; whereas ARID1A deficiency significantly diminished the coupled BRG1-RAD21. Interestingly, the tumor-associated genes within the switched compartments were differentially expressed depending upon Arid1a depletion or not. As a consequence of ARID1A deficiency-induced conformational alteration, the dysregulation of some genes such as PMP22 and GSC, promoted the invasion capacity of liver cancer cells. This study provides an insight into liver cancer tumorigenesis and progression related to ARID1A mutations.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Otto JE, Kadoch C. A two-faced mSWI/SNF subunit: dual roles for ARID1A in tumor suppression and oncogenicity in the liver. Cancer Cell. 2017;32:542–3. - PubMed
-
- Huang J, Deng Q, Wang Q, Li K-Y, Dai J-H, Li N, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44:1117–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 15Z103150093/National Natural Science Foundation of China (National Science Foundation of China)
- 82073116/National Natural Science Foundation of China (National Science Foundation of China)
- 81672772/National Natural Science Foundation of China (National Science Foundation of China)
- 20ZR1427200/Natural Science Foundation of Shanghai (Natural Science Foundation of Shanghai Municipality)
- 19140902500/Natural Science Foundation of Shanghai (Natural Science Foundation of Shanghai Municipality)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

