Genome-wide Gene-by-Sex Interaction Studies Identify Novel Nonsyndromic Orofacial Clefts Risk Locus
- PMID: 34689653
- PMCID: PMC8935575
- DOI: 10.1177/00220345211046614
Genome-wide Gene-by-Sex Interaction Studies Identify Novel Nonsyndromic Orofacial Clefts Risk Locus
Abstract
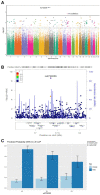
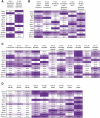
Risk loci identified through genome-wide association studies have explained about 25% of the phenotypic variations in nonsyndromic orofacial clefts (nsOFCs) on the liability scale. Despite the notable sex differences in the incidences of the different cleft types, investigation of loci for sex-specific effects has been understudied. To explore the sex-specific effects in genetic etiology of nsOFCs, we conducted a genome-wide gene × sex (GxSex) interaction study in a sub-Saharan African orofacial cleft cohort. The sample included 1,019 nonsyndromic orofacial cleft cases (814 cleft lip with or without cleft palate and 205 cleft palate only) and 2,159 controls recruited from 3 sites (Ethiopia, Ghana, and Nigeria). An additive logistic model was used to examine the joint effects of the genotype and GxSex interaction. Furthermore, we examined loci with suggestive significance (P < 1E-5) in the additive model for the effect of the GxSex interaction only. We identified a novel risk locus on chromosome 8p22 with genome-wide significant joint and GxSex interaction effects (rs2720555, p2df = 1.16E-08, pGxSex = 1.49E-09, odds ratio [OR] = 0.44, 95% CI = 0.34 to 0.57). For males, the risk of cleft lip with or without cleft palate at this locus decreases with additional copies of the minor allele (p < 0.0001, OR = 0.60, 95% CI = 0.48 to 0.74), but the effect is reversed for females (p = 0.0004, OR = 1.36, 95% CI = 1.15 to 1.60). We replicated the female-specific effect of this locus in an independent cohort (p = 0.037, OR = 1.30, 95% CI = 1.02 to 1.65), but no significant effect was found for the males (p = 0.29, OR = 0.86, 95% CI = 0.65 to 1.14). This locus is in topologically associating domain with craniofacially expressed and enriched genes during embryonic development. Rare coding mutations of some of these genes were identified in nsOFC cohorts through whole exome sequencing analysis. Our study is additional proof that genome-wide GxSex interaction analysis provides an opportunity for novel findings of loci and genes that contribute to the risk of nsOFCs.
Keywords: bioinformatics; craniofacial anomalies; craniofacial biology/genetics; developmental biology; gender differences; gene expression.
Conflict of interest statement
Figures

References
-
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, Baluardo C, Ferrian M, de Assis NA, Alblas MA. 2009. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 41(4):473–477. - PubMed
-
- Burdi AR, Silvey RG. 1969. Sexual differences in closure of the human palatal shelves. Cleft Palate J. 6:1–7. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

