Ammonia Oxidizing Prokaryotes Respond Differently to Fertilization and Termination Methods in Common Oat's Rhizosphere
- PMID: 34690996
- PMCID: PMC8527175
- DOI: 10.3389/fmicb.2021.746524
Ammonia Oxidizing Prokaryotes Respond Differently to Fertilization and Termination Methods in Common Oat's Rhizosphere
Abstract
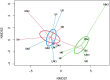
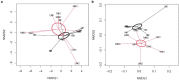
Cover crops (CC) have demonstrated beneficial effects on several soil properties yet questions remain regarding their effects on soil microbial communities. Among them, ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA) have a key role for N cycling in soil and their responses in the rhizosphere of terminated CC deserve further investigation. A greenhouse experiment was established to assess N fertilization (with or without N) and termination methods (glyphosate, mowing, and untreated control) of common oat (Avena sativa L.) as potential drivers of AOA and AOB responses in the rhizosphere. The abundance of amoA genes was determined by quantitative real-time PCR (qPCR), the community structure was assessed with Illumina amplicon sequencing of these genes, while the function was assessed from potential nitrification activity (PNA). While N fertilization had no influence on AOA, the termination method significantly increased amoA gene copies of AOA in mowed plants relative to glyphosate termination or the untreated control (1.76 and 1.49-fold change, respectively), and shifted AOA community structure (PERMANOVA, p<0.05). Ordination methods indicated a separation between AOA communities from control and glyphosate-terminated plants relative to mowed plants for both UniFrac and Aitchison distance. Converserly, N fertilization significantly increased AOB abundance in the rhizosphere of mowed and control plants, yet not in glyphosate-treated plants. Analyses of community structure showed that AOB changed only in response to N fertilization and not to the termination method. In line with these results, significantly higher PNA values were measured in all fertilized samples, regardless of the termination methods. Overall, the results of this study indicated that bacterial and archaeal nitrifiers have contrasting responses to fertlization and plant termination methods. While AOA were responsive to the termination method, AOB were more sensitive to N additions, although, the stimulative effect of N fertilization on amoA AOB abundance was dependent on the termination method.
Keywords: Avena sativa L; amoA; cover crop management; glyphosate; inorganic nitrogen.
Copyright © 2021 Allegrini, Morales, Villamil and Zabaloy.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Aitchison J. The Statistical Analysis of Compositional Data. (1986). London-New York: Chapman and Hall.
-
- Ai C., Liang G., Sun J., Wang X., He P., Zhou W. (2013). Different roles of rhizosphere effect and long-term fertilization in the activity and community structure of ammonia oxidizers in a calcareous fluvo-aquic soil. Soil Biol. Biochem. 57, 30–42. doi: 10.1016/j.soilbio.2012.08.003 - DOI
-
- Aldana A., Carioni D.. (2001). Influencia de la posición topográfica sobre propiedades edáficas de suelos en el predio de la UNS en Altos del Palihue. Trabajo final de tesina. Departamento de Agronomía, Universidad Nacional del Sur. Available at: https://catalogoagronomia.uns.edu.ar (Accessed January 13, 2021).
-
- Allegrini M., Gomez E. D. V., Smalla K., Zabaloy M. C. (2019). Suppression treatment differentially influences the microbial community and the occurrence of broad host range plasmids in the rhizosphere of the model cover crop Avena sativa L. PLoS One 14:e0223600. doi: 10.1371/journal.pone.0223600, PMID: - DOI - PMC - PubMed
-
- Anderson M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x - DOI
LinkOut - more resources
Full Text Sources

