Enzyme Complexes of Ptr4CL and PtrHCT Modulate Co-enzyme A Ligation of Hydroxycinnamic Acids for Monolignol Biosynthesis in Populus trichocarpa
- PMID: 34691108
- PMCID: PMC8527181
- DOI: 10.3389/fpls.2021.727932
Enzyme Complexes of Ptr4CL and PtrHCT Modulate Co-enzyme A Ligation of Hydroxycinnamic Acids for Monolignol Biosynthesis in Populus trichocarpa
Abstract
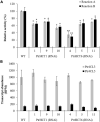
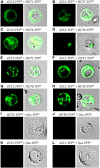
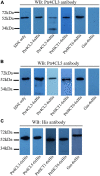
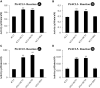
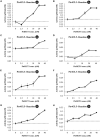
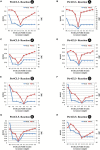
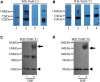
Co-enzyme A (CoA) ligation of hydroxycinnamic acids by 4-coumaric acid:CoA ligase (4CL) is a critical step in the biosynthesis of monolignols. Perturbation of 4CL activity significantly impacts the lignin content of diverse plant species. In Populus trichocarpa, two well-studied xylem-specific Ptr4CLs (Ptr4CL3 and Ptr4CL5) catalyze the CoA ligation of 4-coumaric acid to 4-coumaroyl-CoA and caffeic acid to caffeoyl-CoA. Subsequently, two 4-hydroxycinnamoyl-CoA:shikimic acid hydroxycinnamoyl transferases (PtrHCT1 and PtrHCT6) mediate the conversion of 4-coumaroyl-CoA to caffeoyl-CoA. Here, we show that the CoA ligation of 4-coumaric and caffeic acids is modulated by Ptr4CL/PtrHCT protein complexes. Downregulation of PtrHCTs reduced Ptr4CL activities in the stem-differentiating xylem (SDX) of transgenic P. trichocarpa. The Ptr4CL/PtrHCT interactions were then validated in vivo using biomolecular fluorescence complementation (BiFC) and protein pull-down assays in P. trichocarpa SDX extracts. Enzyme activity assays using recombinant proteins of Ptr4CL and PtrHCT showed elevated CoA ligation activity for Ptr4CL when supplemented with PtrHCT. Numerical analyses based on an evolutionary computation of the CoA ligation activity estimated the stoichiometry of the protein complex to consist of one Ptr4CL and two PtrHCTs, which was experimentally confirmed by chemical cross-linking using SDX plant protein extracts and recombinant proteins. Based on these results, we propose that Ptr4CL/PtrHCT complexes modulate the metabolic flux of CoA ligation for monolignol biosynthesis during wood formation in P. trichocarpa.
Keywords: BiFC; Populus trichocarpa; metabolic flux; monolignol biosynthesis; protein interaction; wood formation.
Copyright © 2021 Lin, Sun, Song, Chen, Shi, Yang, Liu, Tunlaya-Anukit, Liu, Loziuk, Williams, Muddiman, Lin, Sederoff, Wang and Chiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Albersheim P., Darvill A., Roberts K., Sederoff R., Staehelin A. (2010). “The structural polysaccharides of the cell wall and how they are studied,” in Plant Cell Walls, (New York, NY: Taylor & Francis Group; ), 61–84. 10.1201/9780203833476-6 - DOI
-
- Barrière Y., Riboulet C., Méchin V., Maltese S., Pichon M., Cardinal A., et al. (2007). Genetics and genomics of lignification in grass cell walls based on maize as model species. Genes Genomes Genomics 1 133–156.
LinkOut - more resources
Full Text Sources

