Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication
- PMID: 34691511
- PMCID: PMC8290959
- DOI: 10.1093/nsr/nwaa110
Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication
Abstract
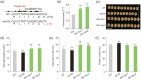
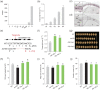
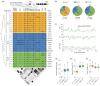
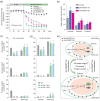
Soybean accounts for more than half of the global production of oilseed and more than a quarter of the protein used globally for human food and animal feed. Soybean domestication involved parallel increases in seed size and oil content, and a concomitant decrease in protein content. However, science has not yet discovered whether these effects were due to selective pressure on a single gene or multiple genes. Here, re-sequencing data from >800 genotypes revealed a strong selection during soybean domestication on GmSWEET10a. The selection of GmSWEET10a conferred simultaneous increases in soybean-seed size and oil content as well as a reduction in the protein content. The result was validated using both near-isogenic lines carrying substitution of haplotype chromosomal segments and transgenic soybeans. Moreover, GmSWEET10b was found to be functionally redundant with its homologue GmSWEET10a and to be undergoing selection in current breeding, leading the the elite allele GmSWEET10b, a potential target for present-day soybean breeding. Both GmSWEET10a and GmSWEET10b were shown to transport sucrose and hexose, contributing to sugar allocation from seed coat to embryo, which consequently determines oil and protein contents and seed size in soybean. We conclude that past selection of optimal GmSWEET10a alleles drove the initial domestication of multiple soybean-seed traits and that targeted selection of the elite allele GmSWEET10b may further improve the yield and seed quality of modern soybean cultivars.
Keywords: SWEET; domestication; seed quality; soybean; yield.
© The Author(s) 2020. Published by Oxford University Press on behalf of China Science Publishing & Media Ltd.
Figures


References
-
- Godfray HCJ, Beddington JR, Crute IR et al. . Food security: the challenge of feeding 9 billion people. Science 2010; 327: 812–8. - PubMed
-
- Foley JA, Ramankutty N, Brauman KA et al. . Solutions for a cultivated planet. Nature 2011; 478: 337–42. - PubMed
-
- Wilson RF. Soybean: market driven research needs. In: Stacey G (ed.). Genetics and Genomics of Soybean. New York: Springer, 2008.
LinkOut - more resources
Full Text Sources
Miscellaneous
