Convergent genomic signatures of high-altitude adaptation among domestic mammals
- PMID: 34692117
- PMCID: PMC8288980
- DOI: 10.1093/nsr/nwz213
Convergent genomic signatures of high-altitude adaptation among domestic mammals
Abstract
Abundant and diverse domestic mammals living on the Tibetan Plateau provide useful materials for investigating adaptive evolution and genetic convergence. Here, we used 327 genomes from horses, sheep, goats, cattle, pigs and dogs living at both high and low altitudes, including 73 genomes generated for this study, to disentangle the genetic mechanisms underlying local adaptation of domestic mammals. Although molecular convergence is comparatively rare at the DNA sequence level, we found convergent signature of positive selection at the gene level, particularly the EPAS1 gene in these Tibetan domestic mammals. We also reported a potential function in response to hypoxia for the gene C10orf67, which underwent positive selection in three of the domestic mammals. Our data provide an insight into adaptive evolution of high-altitude domestic mammals, and should facilitate the search for additional novel genes involved in the hypoxia response pathway.
Keywords: Tibetan Plateau; convergent evolution; domestic animals; genome; high altitude.
© The Author(s) 2020. Published by Oxford University Press on behalf of China Science Publishing & Media Ltd.
Figures
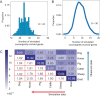
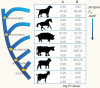

References
-
- Sackton TB, Grayson P, Cloutier Aet al. . Convergent regulatory evolution and loss of flight in paleognathous birds. Science 2019; 364: 74–8. - PubMed
LinkOut - more resources
Full Text Sources
