Robust Glycogene-Based Prognostic Signature for Proficient Mismatch Repair Colorectal Adenocarcinoma
- PMID: 34692502
- PMCID: PMC8529276
- DOI: 10.3389/fonc.2021.727752
Robust Glycogene-Based Prognostic Signature for Proficient Mismatch Repair Colorectal Adenocarcinoma
Abstract
Background: Proficient mismatch repair (pMMR) colorectal adenocarcinoma (CRAC) metastasizes to a greater extent than MMR-deficient CRAC. Prognostic biomarkers are preferred in clinical practice. However, traditional biomarkers screened directly from sequencing are often not robust and thus cannot be confidently utilized.
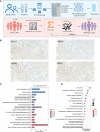
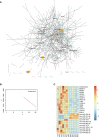
Methods: To circumvent the drawbacks of blind screening, we established a new strategy to identify prognostic biomarkers in the conserved and specific oncogenic pathway and its regulatory RNA network. We performed RNA sequencing (RNA-seq) for messenger RNA (mRNA) and noncoding RNA in six pMMR CRAC patients and constructed a glycosylation-related RNA regulatory network. Biomarkers were selected based on the network and their correlation with the clinicopathologic information and were validated in multiple centers (n = 775).
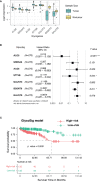
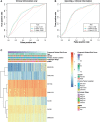
Results: We constructed a competing endogenous RNA (ceRNA) regulatory network using RNA-seq. Genes associated with glycosylation pathways were embedded within this scale-free network. Moreover, we further developed and validated a seven-glycogene prognosis signature, GlycoSig (B3GNT6, GALNT3, GALNT8, ALG8, STT3B, SRD5A3, and ALG6) that prognosticate poor-prognostic subtype for pMMR CRAC patients. This biomarker set was validated in multicenter datasets, demonstrating its robustness and wide applicability. We constructed a simple-to-use nomogram that integrated the risk score of GlycoSig and clinicopathological features of pMMR CRAC patients.
Conclusions: The seven-glycogene signature served as a novel and robust prognostic biomarker set for pMMR CRAC, highlighting the role of a dysregulated glycosylation network in poor prognosis.
Keywords: biomarker; ceRNA; colorectal cancer; glycogene; glycosylation; mismatch repair.
Copyright © 2021 Li, Li, Chen, Lu, Zhou, Li, Zeng, Cai, Lin, Li, Ye, Dong, Yin, Tang, Zhang and Dai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Glycosyltransferase Gene Expression Identifies a Poor Prognostic Colorectal Cancer Subtype Associated with Mismatch Repair Deficiency and Incomplete Glycan Synthesis.Clin Cancer Res. 2018 Sep 15;24(18):4468-4481. doi: 10.1158/1078-0432.CCR-17-3533. Epub 2018 May 29. Clin Cancer Res. 2018. PMID: 29844132
-
Combination of lysine-specific demethylase 6A (KDM6A) and mismatch repair (MMR) status is a potential prognostic factor in colorectal cancer.Cancer Med. 2021 Jan;10(1):317-324. doi: 10.1002/cam4.3602. Epub 2020 Nov 11. Cancer Med. 2021. PMID: 33174323 Free PMC article.
-
Lymphocytic response to tumour and deficient DNA mismatch repair identify subtypes of stage II/III colorectal cancer associated with patient outcomes.Gut. 2019 Mar;68(3):465-474. doi: 10.1136/gutjnl-2017-315664. Epub 2018 Jan 30. Gut. 2019. PMID: 29382774
-
Analysis of Circular RNA-Related Competing Endogenous RNA Identifies the Immune-Related Risk Signature for Colorectal Cancer.Front Genet. 2020 Jun 3;11:505. doi: 10.3389/fgene.2020.00505. eCollection 2020. Front Genet. 2020. PMID: 32582276 Free PMC article.
-
Weighted Gene Co-expression Network Analysis Identifies CALD1 as a Biomarker Related to M2 Macrophages Infiltration in Stage III and IV Mismatch Repair-Proficient Colorectal Carcinoma.Front Mol Biosci. 2021 Apr 29;8:649363. doi: 10.3389/fmolb.2021.649363. eCollection 2021. Front Mol Biosci. 2021. PMID: 33996905 Free PMC article.
Cited by
-
Glycosyltransferases in Cancer: Prognostic Biomarkers of Survival in Patient Cohorts and Impact on Malignancy in Experimental Models.Cancers (Basel). 2022 Apr 24;14(9):2128. doi: 10.3390/cancers14092128. Cancers (Basel). 2022. PMID: 35565254 Free PMC article. Review.
-
Glycosylation-Related Genes Predict the Prognosis and Immune Fraction of Ovarian Cancer Patients Based on Weighted Gene Coexpression Network Analysis (WGCNA) and Machine Learning.Oxid Med Cell Longev. 2022 Mar 4;2022:3665617. doi: 10.1155/2022/3665617. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35281472 Free PMC article.
References
LinkOut - more resources
Full Text Sources

