Dietary Energy Level Impacts the Performance of Donkeys by Manipulating the Gut Microbiome and Metabolome
- PMID: 34692802
- PMCID: PMC8531409
- DOI: 10.3389/fvets.2021.694357
Dietary Energy Level Impacts the Performance of Donkeys by Manipulating the Gut Microbiome and Metabolome
Abstract
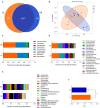
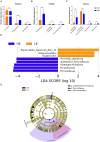
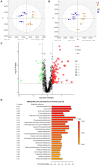
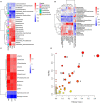
Considerable evidence suggests that dietary energy levels and gut microbiota are pivotal for animal health and productivity. However, little information exists about the correlations among dietary energy level, performance, and the gut microbiota and metabolome of donkeys. The objective of this study was to investigate the mechanisms by which dietary energy content dictates the growth performance by modulating the intestinal microbiome and metabolome of donkeys. Thirty-six nine-month-old male Dezhou donkeys with similar body weights were randomly assigned to two groups fed low- or high-energy diets (LE or HE). The results showed that donkeys fed HE had increased (p < 0.05) the average daily gain (ADG) and feed efficiency (G/F) compared with those that received LE diet. The gut microbiota in both groups was dominated by the phyla Firmicutes and Bacteroidetes regardless of the dietary energy level. However, feeding HE to donkeys significantly decreased (p < 0.05) the ratio of Firmicutes to Bacteroidetes (F/B). Compared to the LE group, feeding HE specifically increased the abundances of unidentified_Prevotellaceae (p = 0.02) while decreasing the richness of unidentified_Ruminococcaceae (p = 0.05). Compared to the LE group, feeding the HE diet significantly (p < 0.05) upregulated certain metabolic pathways involving the aspartate metabolism and the urea cycle. In addition, the increased bacteria and metabolites in the HE-fed group exhibited a positive correlation with improved growth performance of donkeys. Taken together, feeding the HE diet increased the richness of Prevotellaceae and upregulated growth-related metabolic pathways, which may have contributed to the ameliorated growth performance of donkeys. Thus, it is a recommendable dietary strategy to feed HE diets to fattening donkeys for superior product performance and feed efficiency.
Keywords: digestible energy; donkey; growth performance; metabolome; microbiome.
Copyright © 2021 Zhang, Zhang, Wang, Du, Zhang and Lee.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources

