Mechanism of activation and the rewired network: New drug design concepts
- PMID: 34693559
- PMCID: PMC8837674
- DOI: 10.1002/med.21863
Mechanism of activation and the rewired network: New drug design concepts
Abstract
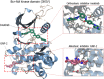
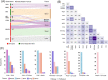
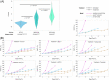
Precision oncology benefits from effective early phase drug discovery decisions. Recently, drugging inactive protein conformations has shown impressive successes, raising the cardinal questions of which targets can profit and what are the principles of the active/inactive protein pharmacology. Cancer driver mutations have been established to mimic the protein activation mechanism. We suggest that the decision whether to target an inactive (or active) conformation should largely rest on the protein mechanism of activation. We next discuss the recent identification of double (multiple) same-allele driver mutations and their impact on cell proliferation and suggest that like single driver mutations, double drivers also mimic the mechanism of activation. We further suggest that the structural perturbations of double (multiple) in cis mutations may reveal new surfaces/pockets for drug design. Finally, we underscore the preeminent role of the cellular network which is deregulated in cancer. Our structure-based review and outlook updates the traditional Mechanism of Action, informs decisions, and calls attention to the intrinsic activation mechanism of the target protein and the rewired tumor-specific network, ushering innovative considerations in precision medicine.
Keywords: K-Ras4B; KRAS; cancer network; driver mutations; drug discovery; inhibitor; kinases.
© 2021 The Authors. Medicinal Research Reviews published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures



Similar articles
-
Why Are Some Driver Mutations Rare?Trends Pharmacol Sci. 2019 Dec;40(12):919-929. doi: 10.1016/j.tips.2019.10.003. Epub 2019 Nov 5. Trends Pharmacol Sci. 2019. PMID: 31699406 Review.
-
Inhibition of Nonfunctional Ras.Cell Chem Biol. 2021 Feb 18;28(2):121-133. doi: 10.1016/j.chembiol.2020.12.012. Epub 2021 Jan 12. Cell Chem Biol. 2021. PMID: 33440168 Free PMC article. Review.
-
Anticancer drug resistance: An update and perspective.Drug Resist Updat. 2021 Dec;59:100796. doi: 10.1016/j.drup.2021.100796. Epub 2021 Dec 16. Drug Resist Updat. 2021. PMID: 34953682 Free PMC article. Review.
-
Precision medicine review: rare driver mutations and their biophysical classification.Biophys Rev. 2019 Feb;11(1):5-19. doi: 10.1007/s12551-018-0496-2. Epub 2019 Jan 4. Biophys Rev. 2019. PMID: 30610579 Free PMC article. Review.
-
Review: Precision medicine and driver mutations: Computational methods, functional assays and conformational principles for interpreting cancer drivers.PLoS Comput Biol. 2019 Mar 28;15(3):e1006658. doi: 10.1371/journal.pcbi.1006658. eCollection 2019 Mar. PLoS Comput Biol. 2019. PMID: 30921324 Free PMC article. Review.
Cited by
-
The mechanism of activation of MEK1 by B-Raf and KSR1.Cell Mol Life Sci. 2022 May 4;79(5):281. doi: 10.1007/s00018-022-04296-0. Cell Mol Life Sci. 2022. PMID: 35508574 Free PMC article.
-
Allosteric regulation of autoinhibition and activation of c-Abl.Comput Struct Biotechnol J. 2022 Aug 11;20:4257-4270. doi: 10.1016/j.csbj.2022.08.014. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36051879 Free PMC article.
-
Regulatory spine RS3 residue of protein kinases: a lipophilic bystander or a decisive element in the small-molecule kinase inhibitor binding?Biochem Soc Trans. 2022 Feb 28;50(1):633-648. doi: 10.1042/BST20210837. Biochem Soc Trans. 2022. PMID: 35226061 Free PMC article.
-
A New View of Activating Mutations in Cancer.Cancer Res. 2022 Nov 15;82(22):4114-4123. doi: 10.1158/0008-5472.CAN-22-2125. Cancer Res. 2022. PMID: 36069825 Free PMC article.
-
CDK2 and CDK4: Cell Cycle Functions Evolve Distinct, Catalysis-Competent Conformations, Offering Drug Targets.JACS Au. 2024 May 14;4(5):1911-1927. doi: 10.1021/jacsau.4c00138. eCollection 2024 May 27. JACS Au. 2024. PMID: 38818077 Free PMC article.
References
-
- Saito Y, Koya J, Araki M, et al. Landscape and function of multiple mutations within individual oncogenes. Nature. 2020;582(7810):95‐99. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

