Screening of Minimalist Noncanonical Sites in Duplex DNA and RNA Reveals Context and Motif-Selective Binding by Fluorogenic Base Probes
- PMID: 34693570
- PMCID: PMC8758549
- DOI: 10.1002/chem.202103616
Screening of Minimalist Noncanonical Sites in Duplex DNA and RNA Reveals Context and Motif-Selective Binding by Fluorogenic Base Probes
Abstract
We hypothesize that programmable hybridization to noncanonical nucleic acid motifs may be achieved by macromolecular display of binders to individual noncanonical pairs (NCPs). As each recognition element may individually have weak binding to an NCP, we developed a semi-rational approach to detect low affinity interactions between selected nitrogenous bases and noncanonical sites in duplex DNA and RNA. A set of fluorogenic probes was synthesized by coupling abiotic (triazines, pyrimidines) and native RNA bases to thiazole orange (TO) dye. This probe library was screened against duplex nucleic acid substrates bearing single abasic, single NCP, and tandem NCP sites. Probe engagement with NCP sites was reported by 100-1000× fluorescence enhancement over background. Binding is strongly context-dependent, reflective of both molecular recognition and stability: less stable motifs are more likely to bind a synthetic probe. Further, DNA and RNA substrates exhibit entirely different abasic and single NCP binding profiles. While probe binding in the abasic and single NCP screens was monotonous, much richer binding profiles were observed with the screen of tandem NCP sites in RNA, in part due to increased steric accessibility. In addition to known binding interactions between the triazine melamine (M) and T/U sites, the NCP screens identified new targeting elements for pyrimidine-rich motifs in single NCPs and 2×2 internal bulges. We anticipate that semi-rational approaches of this type will lead to programmable noncanonical hybridization strategies at the macromolecular level.
Keywords: RNA recognition; fluorescent probes; noncanonical pairs; nucleobases; synthetic bases.
© 2021 Wiley-VCH GmbH.
Conflict of interest statement
Conflict of interest
The authors declare no conflict of interest.
Figures




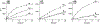
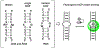
Similar articles
-
DNA Stains as Surrogate Nucleobases in Fluorogenic Hybridization Probes.Acc Chem Res. 2016 Apr 19;49(4):714-23. doi: 10.1021/acs.accounts.5b00546. Epub 2016 Mar 10. Acc Chem Res. 2016. PMID: 26963493
-
Single labeled DNA FIT probes for avoiding false-positive signaling in the detection of DNA/RNA in qPCR or cell media.Chembiochem. 2012 Sep 24;13(14):2072-81. doi: 10.1002/cbic.201200397. Epub 2012 Aug 31. Chembiochem. 2012. PMID: 22936610
-
Purine-Derived Nitroxides for Noncovalent Spin-Labeling of Abasic Sites in Duplex Nucleic Acids.Chemistry. 2018 Mar 15;24(16):4157-4164. doi: 10.1002/chem.201705410. Epub 2018 Feb 16. Chemistry. 2018. PMID: 29451325
-
DNA probes: applications of the principles of nucleic acid hybridization.Crit Rev Biochem Mol Biol. 1991;26(3-4):227-59. doi: 10.3109/10409239109114069. Crit Rev Biochem Mol Biol. 1991. PMID: 1718662 Review.
-
Nucleic acid-based fluorescent probes and their analytical potential.Anal Bioanal Chem. 2011 Mar;399(9):3157-76. doi: 10.1007/s00216-010-4304-5. Epub 2010 Oct 29. Anal Bioanal Chem. 2011. PMID: 21046088 Free PMC article. Review.
Cited by
-
Intracellular RNA and DNA tracking by uridine-rich internal loop tagging with fluorogenic bPNA.Nat Commun. 2023 May 24;14(1):2987. doi: 10.1038/s41467-023-38579-2. Nat Commun. 2023. PMID: 37225690 Free PMC article.
-
Synthesis of Bifacial Peptide Nucleic Acids with Diketopiperazine Backbones.Synlett. 2022 Jun;33(10):965-968. doi: 10.1055/a-1802-6873. Epub 2022 Apr 28. Synlett. 2022. PMID: 35874045 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

