Fungal lysozyme leverages the gut microbiota to curb DSS-induced colitis
- PMID: 34693864
- PMCID: PMC8547870
- DOI: 10.1080/19490976.2021.1988836
Fungal lysozyme leverages the gut microbiota to curb DSS-induced colitis
Abstract
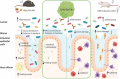
Colitis is characterized by colonic inflammation and impaired gut health. Both features aggravate obesity and insulin resistance. Host defense peptides (HDPs) are key regulators of gut homeostasis and generally malfunctioning in above-mentioned conditions. We aimed here to improve bowel function in diet-induced obesity and chemically induced colitis through daily oral administration of lysozyme, a well-characterized HDP, derived from Acremonium alcalophilum.C57BL6/J mice were fed either low-fat reference diet or HFD ± daily gavage of lysozyme for 12 weeks, followed by metabolic assessment and evaluation of colonic microbiota encroachment. To further evaluate the efficacy of intestinal inflammation, we next supplemented chow-fed BALB/c mice with lysozyme during Dextran Sulfate Sodium (DSS)-induced colitis in either conventional or microbiota-depleted mice. We assessed longitudinal microbiome alterations by 16S amplicon sequencing in both models.Lysozyme dose-dependently alleviated intestinal inflammation in DSS-challenged mice and further protected against HFD-induced microbiota encroachment and fasting hyperinsulinemia. Observed improvements of intestinal health relied on a complex gut flora, with the observation that microbiota depletion abrogated lysozyme's capacity to mitigate DSS-induced colitis.Akkermansia muciniphila associated with impaired gut health in both models, a trajectory that was mitigated by lysozyme administration. In agreement with this notion, PICRUSt2 analysis revealed specific pathways consistently affected by lysozyme administration, independent of vivarium, disease model and mouse strain.Taking together, lysozyme leveraged the gut microbiota to curb DSS-induced inflammation, alleviated HFD-induced gastrointestinal disturbances and lowered fasting insulin levels in obese mice. Collectively, these data present A. alcalophilum-derived lysozyme as a promising candidate to enhance gut health.
Keywords: Gut health; colitis; high fat diet; host defense peptides; insulin resistance; intestinal inflammation; microbiota encroachment; microbiota function; mucus; muramidase.
Conflict of interest statement
NNK, EGWS, PBO and LBSH are employed at Novozymes A/S (NZAS). NZAS provided the test compound and filed a patent based on the enclosed findings (PCT/EP2018/050189). The views presented in this manuscript are those of the authors and not necessarily those of NZAS.
Figures





References
-
- Volynets V, Louis S, Pretz D, Lang L, Ostaff MJ, Wehkamp J, Bischoff SC.. Intestinal barrier function and the gut microbiome are differentially affected in mice fed a western-style diet or drinking water supplemented with fructose. J. Nutr. 2017;147:770–20. doi: 10.3945/jn.116.242859. - DOI - PubMed
-
- Lam YY, Ha CWY, Hoffmann JMA, Oscarsson J, Dinudom A, Mather TJ, Cook DI, Hunt NH, Caterson ID, Holmes AJ, et al. Effects of dietary fat profile on gut permeability and microbiota and their relationships with metabolic changes in mice. Obesity. 2015;23(7):1429–1439. doi: 10.1002/oby.21122. - DOI - PubMed
-
- Jensen BAH, Nielsen TS, Fritzen AM, Holm JB, Fjære E, Serup AK, Borkowski K, Risis S, Pærregaard SI, Søgaard I, et al. Dietary fat drives whole-body insulin resistance and promotes intestinal inflammation independent of body weight gain. Metabolism. 2016;65(12):1706–1719. doi: 10.1016/j.metabol.2016.09.002. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
