Cnm1 mediates nucleus-mitochondria contact site formation in response to phospholipid levels
- PMID: 34694322
- PMCID: PMC8548916
- DOI: 10.1083/jcb.202104100
Cnm1 mediates nucleus-mitochondria contact site formation in response to phospholipid levels
Abstract
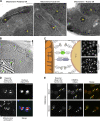
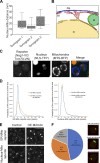
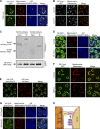
Mitochondrial functions are tightly regulated by nuclear activity, requiring extensive communication between these organelles. One way by which organelles can communicate is through contact sites, areas of close apposition held together by tethering molecules. While many contacts have been characterized in yeast, the contact between the nucleus and mitochondria was not previously identified. Using fluorescence and electron microscopy in S. cerevisiae, we demonstrate specific areas of contact between the two organelles. Using a high-throughput screen, we uncover a role for the uncharacterized protein Ybr063c, which we have named Cnm1 (contact nucleus mitochondria 1), as a molecular tether on the nuclear membrane. We show that Cnm1 mediates contact by interacting with Tom70 on mitochondria. Moreover, Cnm1 abundance is regulated by phosphatidylcholine, enabling the coupling of phospholipid homeostasis with contact extent. The discovery of a molecular mechanism that allows mitochondrial crosstalk with the nucleus sets the ground for better understanding of mitochondrial functions in health and disease.
© 2021 Eisenberg-Bord et al.
Figures








Comment in
-
Cnm1: A bridge between mitochondria and nuclear ER.J Cell Biol. 2021 Nov 1;220(11):e202109021. doi: 10.1083/jcb.202109021. Epub 2021 Oct 25. J Cell Biol. 2021. PMID: 34694321 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

