Visualization and Quantification of Subcellular RNA Localization Using Single-Molecule RNA Fluorescence In Situ Hybridization
- PMID: 34694613
- PMCID: PMC9038117
- DOI: 10.1007/978-1-0716-1851-6_13
Visualization and Quantification of Subcellular RNA Localization Using Single-Molecule RNA Fluorescence In Situ Hybridization
Abstract
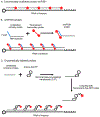
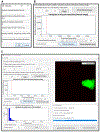
Advancements in imaging technologies, especially approaches that allow the imaging of single RNA molecules, have opened new avenues to understand RNA regulation, from synthesis to decay with high spatial and temporal resolution. Here, we describe a protocol for single-molecule fluorescent in situ hybridization (smFISH) using three different approaches for synthesizing the fluorescent probes. The three approaches described are commercially available probes, single-molecule inexpensive FISH (smiFISH), and in-house enzymatically labeled probes. These approaches offer technical and economic flexibility to meet the specific needs of an experiment. In addition, we provide a protocol to perform automated smFISH spot detection using the software FISH-quant.
Keywords: Fluorescence in situ hybridization; Fluorescence microscopy; RNA imaging; RNA localization; RNA transport; Single-molecule RNA quantification.
© 2022. The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature.
Figures








References
-
- Kim-Ha J, Smith JL, and Macdonald PM (1991) oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 66:23–35 - PubMed
-
- St Johnston D, Beuchle D, and Nüsslein-Volhard C (1991) Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 66:51–63 - PubMed
-
- Mowry KL and Melton DA (1992) Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science 255:991–994 - PubMed
-
- Long RM, Singer RH, Meng X, et al. (1997) Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277:383–387 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

