S-farnesylation is essential for antiviral activity of the long ZAP isoform against RNA viruses with diverse replication strategies
- PMID: 34695163
- PMCID: PMC8568172
- DOI: 10.1371/journal.ppat.1009726
S-farnesylation is essential for antiviral activity of the long ZAP isoform against RNA viruses with diverse replication strategies
Abstract
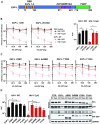
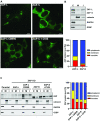
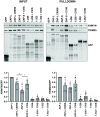
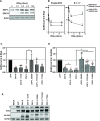
The zinc finger antiviral protein (ZAP) is a broad inhibitor of virus replication. Its best-characterized function is to bind CpG dinucleotides present in viral RNAs and, through the recruitment of TRIM25, KHNYN and other cofactors, target them for degradation or prevent their translation. The long and short isoforms of ZAP (ZAP-L and ZAP-S) have different intracellular localization and it is unclear how this regulates their antiviral activity against viruses with different sites of replication. Using ZAP-sensitive and ZAP-insensitive human immunodeficiency virus type I (HIV-1), which transcribe the viral RNA in the nucleus and assemble virions at the plasma membrane, we show that the catalytically inactive poly-ADP-ribose polymerase (PARP) domain in ZAP-L is essential for CpG-specific viral restriction. Mutation of a crucial cysteine in the C-terminal CaaX box that mediates S-farnesylation and, to a lesser extent, the residues in place of the catalytic site triad within the PARP domain, disrupted the activity of ZAP-L. Addition of the CaaX box to ZAP-S partly restored antiviral activity, explaining why ZAP-S lacks antiviral activity for CpG-enriched HIV-1 despite conservation of the RNA-binding domain. Confocal microscopy confirmed the CaaX motif mediated localization of ZAP-L to vesicular structures and enhanced physical association with intracellular membranes. Importantly, the PARP domain and CaaX box together jointly modulate the interaction between ZAP-L and its cofactors TRIM25 and KHNYN, implying that its proper subcellular localisation is required to establish an antiviral complex. The essential contribution of the PARP domain and CaaX box to ZAP-L antiviral activity was further confirmed by inhibition of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replication, which replicates in double-membrane vesicles derived from the endoplasmic reticulum. Thus, compartmentalization of ZAP-L on intracellular membranes provides an essential effector function in ZAP-L-mediated antiviral activity against divergent viruses with different subcellular replication sites.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

