Single-Cell Transcriptomics Reveals a Conserved Metaplasia Program in Pancreatic Injury
- PMID: 34695382
- PMCID: PMC8792222
- DOI: 10.1053/j.gastro.2021.10.027
Single-Cell Transcriptomics Reveals a Conserved Metaplasia Program in Pancreatic Injury
Abstract
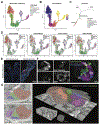
Background & aims: Acinar to ductal metaplasia (ADM) occurs in the pancreas in response to tissue injury and is a potential precursor for adenocarcinoma. The goal of these studies was to define the populations arising from ADM, the associated transcriptional changes, and markers of disease progression.
Methods: Acinar cells were lineage-traced with enhanced yellow fluorescent protein (EYFP) to follow their fate post-injury. Transcripts of more than 13,000 EYFP+ cells were determined using single-cell RNA sequencing (scRNA-seq). Developmental trajectories were generated. Data were compared with gastric metaplasia, KrasG12D-induced neoplasia, and human pancreatitis. Results were confirmed by immunostaining and electron microscopy. KrasG12D was expressed in injury-induced ADM using several inducible Cre drivers. Surgical specimens of chronic pancreatitis from 15 patients were evaluated by immunostaining.
Results: scRNA-seq of ADM revealed emergence of a mucin/ductal population resembling gastric pyloric metaplasia. Lineage trajectories suggest that some pyloric metaplasia cells can generate tuft and enteroendocrine cells (EECs). Comparison with KrasG12D-induced ADM identifies populations associated with disease progression. Activation of KrasG12D expression in HNF1B+ or POU2F3+ ADM populations leads to neoplastic transformation and formation of MUC5AC+ gastric-pit-like cells. Human pancreatitis samples also harbor pyloric metaplasia with a similar transcriptional phenotype.
Conclusions: Under conditions of chronic injury, acinar cells undergo a pyloric-type metaplasia to mucinous progenitor-like populations, which seed disparate tuft cell and EEC lineages. ADM-derived EEC subtypes are diverse. KrasG12D expression is sufficient to drive neoplasia when targeted to injury-induced ADM populations and offers an alternative origin for tumorigenesis. This program is conserved in human pancreatitis, providing insight into early events in pancreas diseases.
Keywords: ADM; Enteroendocrine Cells; Paligenosis; Plasticity; Tuft Cells.
Copyright © 2022 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
- T32 LM012412/LM/NLM NIH HHS/United States
- R01 DK101332/DK/NIDDK NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- R21 DK111369/DK/NIDDK NIH HHS/United States
- T32 CA009547/CA/NCI NIH HHS/United States
- T32 CA009370/CA/NCI NIH HHS/United States
- I01 BX000930/BX/BLRD VA/United States
- T32 CA119925/CA/NCI NIH HHS/United States
- R01 CA246208/CA/NCI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- R01 DK110406/DK/NIDDK NIH HHS/United States
- R01 DK103831/DK/NIDDK NIH HHS/United States
- R01 CA239645/CA/NCI NIH HHS/United States
- R01 DK094989/DK/NIDDK NIH HHS/United States
- U01 CA215798/CA/NCI NIH HHS/United States
- R35 GM142709/GM/NIGMS NIH HHS/United States
- 27145/CRUK_/Cancer Research UK/United Kingdom
- P30 DK056338/DK/NIDDK NIH HHS/United States
- P41 GM103712/GM/NIGMS NIH HHS/United States
- P50 CA236733/CA/NCI NIH HHS/United States
- U01 DK120447/DK/NIDDK NIH HHS/United States
- R35 CA197687/CA/NCI NIH HHS/United States
- R03 DK127484/DK/NIDDK NIH HHS/United States
- R01 DK105129/DK/NIDDK NIH HHS/United States
- P30 CA014195/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

