Evolutionary Dynamics and Epidemiology of Endemic and Emerging Coronaviruses in Humans, Domestic Animals, and Wildlife
- PMID: 34696338
- PMCID: PMC8537103
- DOI: 10.3390/v13101908
Evolutionary Dynamics and Epidemiology of Endemic and Emerging Coronaviruses in Humans, Domestic Animals, and Wildlife
Abstract
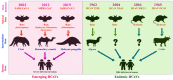
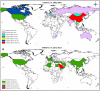
Diverse coronavirus (CoV) strains can infect both humans and animals and produce various diseases. CoVs have caused three epidemics and pandemics in the last two decades, and caused a severe impact on public health and the global economy. Therefore, it is of utmost importance to understand the emergence and evolution of endemic and emerging CoV diversity in humans and animals. For diverse bird species, the Infectious Bronchitis Virus is a significant one, whereas feline enteric and canine coronavirus, recombined to produce feline infectious peritonitis virus, infects wild cats. Bovine and canine CoVs have ancestral relationships, while porcine CoVs, especially SADS-CoV, can cross species barriers. Bats are considered as the natural host of diverse strains of alpha and beta coronaviruses. Though MERS-CoV is significant for both camels and humans, humans are nonetheless affected more severely. MERS-CoV cases have been reported mainly in the Arabic peninsula since 2012. To date, seven CoV strains have infected humans, all descended from animals. The severe acute respiratory syndrome coronaviruses (SARS-CoV and SARS-CoV-2) are presumed to be originated in Rhinolopoid bats that severely infect humans with spillover to multiple domestic and wild animals. Emerging alpha and delta variants of SARS-CoV-2 were detected in pets and wild animals. Still, the intermediate hosts and all susceptible animal species remain unknown. SARS-CoV-2 might not be the last CoV to cross the species barrier. Hence, we recommend developing a universal CoV vaccine for humans so that any future outbreak can be prevented effectively. Furthermore, a One Health approach coronavirus surveillance should be implemented at human-animal interfaces to detect novel coronaviruses before emerging to humans and to prevent future epidemics and pandemics.
Keywords: MERS-CoV; Rhinolopoid; SARS-CoV; SARS-CoV-2; bats; surveillance; zoonotic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


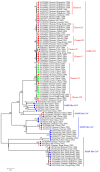
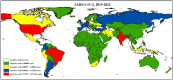
References
-
- Islam A., Sayeed M.A., Rahman M.K., Ferdous J., Shano S., Choudhury S.D., Hassan M.M. Spatiotemporal patterns and trends of community transmission of the pandemic COVID-19 in South Asia: Bangladesh as a case study. Biosaf. Health. 2020;3:39–49. doi: 10.1016/j.bsheal.2020.09.006. - DOI - PMC - PubMed
-
- WHO Virtual Press Conference on COVID-19—11 March 2020. 2020. [(accessed on 8 August 2021)]. Available online: https://www.who.int/docs/default-source/coronaviruse/transcripts/who-aud....
-
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

