SARS-CoV-2 Subgenomic RNAs: Characterization, Utility, and Perspectives
- PMID: 34696353
- PMCID: PMC8539008
- DOI: 10.3390/v13101923
SARS-CoV-2 Subgenomic RNAs: Characterization, Utility, and Perspectives
Erratum in
-
Correction: Long, S. SARS-CoV-2 Subgenomic RNAs: Characterization, Utility, and Perspectives. Viruses 2020, 13, 1923.Viruses. 2022 Jun 28;14(7):1406. doi: 10.3390/v14071406. Viruses. 2022. PMID: 35891574 Free PMC article.
Abstract
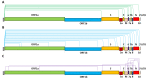
SARS-CoV-2, the etiologic agent at the root of the ongoing COVID-19 pandemic, harbors a large RNA genome from which a tiered ensemble of subgenomic RNAs (sgRNAs) is generated. Comprehensive definition and investigation of these RNA products are important for understanding SARS-CoV-2 pathogenesis. This review summarizes the recent progress on SARS-CoV-2 sgRNA identification, characterization, and application as a viral replication marker. The significance of these findings and potential future research areas of interest are discussed.
Keywords: COVID-19; SARS-CoV-2; sgRNA; subgenomic RNA.
Conflict of interest statement
The author declares no known competing financial interests or personal relationships that could have appeared to influence the work reported in this manuscript.
Figures


References
-
- Sawicki S.G., Sawicki D.L. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 1995;380:499–506. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

