Genomic Signatures in HPV-Associated Tumors
- PMID: 34696429
- PMCID: PMC8537705
- DOI: 10.3390/v13101998
Genomic Signatures in HPV-Associated Tumors
Abstract
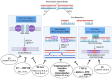
Papillomaviruses dysregulate the G1/S cell cycle transition in order to promote DNA synthesis in S phase, which is a requirement for viral replication. The human papillomaviruses (HPV) E6 and E7 oncoproteins mediate degradation of the cell cycle regulators p53 and Rb, which are two of the most universally disrupted tumor-suppressor genes in all of cancer. The G1/S checkpoint is activated in normal cells to allow sufficient time for DNA repair in G1 before proceeding to replicate DNA and risk propagating unrepaired errors. The TP53 pathway suppresses a variety of such errors, including translocation, copy number alterations, and aneuploidy, which are thus found in HPV-associated tumors similarly to HPV-negative tumors with other mechanisms of TP53 disruption. However, E6 and E7 maintain a variety of other virus-host interactions that directly disrupt a growing list of other DNA repair and chromatin remodeling factors, implying HPV-specific repair deficiencies. In addition, HPV-associated squamous cell carcinomas tumors clinically respond differently to DNA damaging agents compared to their HPV negative counterparts. The focus of this review is to integrate three categories of observations: (1) pre-clinical understanding as to the effect of HPV on DNA repair, (2) genomic signatures of DNA repair in HPV-associated tumor genomes, and (3) clinical responses of HPV-associated tumors to DNA damaging agents. The goals are to try to explain why HPV-associated tumors respond so well to DNA damaging agents, identify missing pieces, and suggest clinical strategies could be used to further improve treatment of these cancers.
Keywords: DNA repair; E6 E7 oncoproteins; alternative end-joining; human papillomavirus.
Conflict of interest statement
J.E.L. and D.S.H. are listed inventors on a patent filed by Memorial Sloan Kettering Cancer Center regarding the use of alternative end-joining genomic signatures to predict radiation sensitivity. There are no licenses or royalties. DSH has a research contract with SQZBiotechnologies for an unrelated project and has received travel funds from Biorad, Inc.
Figures
References
-
- Helen F., Cancer Genome Atlas Research Network. Albert Einstein College of Medicine. Analytical Biological Services. Barretos Cancer Hospital. Baylor College of Medicine. Beckman Research Institute of City of Hope. Buck Institute for Research on Aging. Canada’s Michael Smith Genome Sciences Centre. Harvard Medical School et al. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378–384. doi: 10.1038/nature21386. - DOI - PMC - PubMed
-
- Chung J.H., Sanford E., Johnson A., Klempner S.J., Schrock A.B., Palma N.A., Erlich R.L., Frampton G.M., Chalmers Z.R., Vergilio J., et al. Comprehensive genomic profiling of anal squamous cell carcinoma reveals distinct genomically defined classes. Ann. Oncol. 2016;27:1336–1341. doi: 10.1093/annonc/mdw152. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous