Discovery and Genetic Characterization of Novel Paramyxoviruses Related to the Genus Henipavirus in Crocidura Species in the Republic of Korea
- PMID: 34696450
- PMCID: PMC8537881
- DOI: 10.3390/v13102020
Discovery and Genetic Characterization of Novel Paramyxoviruses Related to the Genus Henipavirus in Crocidura Species in the Republic of Korea
Abstract
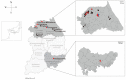
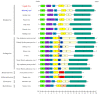
Paramyxoviruses, negative-sense single-stranded RNA viruses, pose a critical threat to human public health. Currently, 78 species, 17 genera, and 4 subfamilies of paramyxoviruses are harbored by multiple natural reservoirs, including rodents, bats, birds, reptiles, and fish. Henipaviruses are critical zoonotic pathogens that cause severe acute respiratory distress and neurological diseases in humans. Using reverse transcription-polymerase chain reaction, 115 Crocidura species individuals were examined for the prevalence of paramyxovirus infections. Paramyxovirus RNA was observed in 26 (22.6%) shrews collected at five trapping sites, Republic of Korea. Herein, we report two genetically distinct novel paramyxoviruses (genus: Henipavirus): Gamak virus (GAKV) and Daeryong virus (DARV) isolated from C. lasiura and C. shantungensis, respectively. Two GAKVs and one DARV were nearly completely sequenced using next-generation sequencing. GAKV and DARV contain six genes (3'-N-P-M-F-G-L-5') with genome sizes of 18,460 nucleotides and 19,471 nucleotides, respectively. The phylogenetic inference demonstrated that GAKV and DARV form independent genetic lineages of Henipavirus in Crocidura species. GAKV-infected human lung epithelial cells elicited the induction of type I/III interferons, interferon-stimulated genes, and proinflammatory cytokines. In conclusion, this study contributes further understandings of the molecular prevalence, genetic characteristics and diversity, and zoonotic potential of novel paramyxoviruses in shrews.
Keywords: Crocidura paramyxovirus; genetic characterization and diversity; next-generation sequencing; novel virus discovery; potential zoonosis.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

