Spike Proteins of SARS-CoV-2 Induce Pathological Changes in Molecular Delivery and Metabolic Function in the Brain Endothelial Cells
- PMID: 34696455
- PMCID: PMC8538996
- DOI: 10.3390/v13102021
Spike Proteins of SARS-CoV-2 Induce Pathological Changes in Molecular Delivery and Metabolic Function in the Brain Endothelial Cells
Abstract
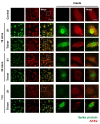
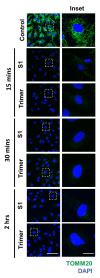
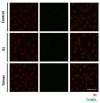
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes the coronavirus disease (COVID-19), is currently infecting millions of people worldwide and is causing drastic changes in people's lives. Recent studies have shown that neurological symptoms are a major issue for people infected with SARS-CoV-2. However, the mechanism through which the pathological effects emerge is still unclear. Brain endothelial cells (ECs), one of the components of the blood-brain barrier, are a major hurdle for the entry of pathogenic or infectious agents into the brain. They strongly express angiotensin converting enzyme 2 (ACE2) for its normal physiological function, which is also well-known to be an opportunistic receptor for SARS-CoV-2 spike protein, facilitating their entry into host cells. First, we identified rapid internalization of the receptor-binding domain (RBD) S1 domain (S1) and active trimer (Trimer) of SARS-CoV-2 spike protein through ACE2 in brain ECs. Moreover, internalized S1 increased Rab5, an early endosomal marker while Trimer decreased Rab5 in the brain ECs. Similarly, the permeability of transferrin and dextran was increased in S1 treatment but decreased in Trimer, respectively. Furthermore, S1 and Trimer both induced mitochondrial damage including functional deficits in mitochondrial respiration. Overall, this study shows that SARS-CoV-2 itself has toxic effects on the brain ECs including defective molecular delivery and metabolic function, suggesting a potential pathological mechanism to induce neurological signs in the brain.
Keywords: BBB; COVID-19; SARS-CoV-2; endothelial cells; metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- WHO Coronavirus Disease (COVID-19) Dashboard: World Health Organization. 2021. [(accessed on 5 October 2021)]. Available online: https://covid19.who.int/
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

