An Update on Innate Immune Responses during SARS-CoV-2 Infection
- PMID: 34696490
- PMCID: PMC8541410
- DOI: 10.3390/v13102060
An Update on Innate Immune Responses during SARS-CoV-2 Infection
Abstract
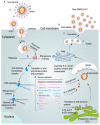
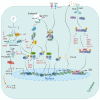
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a member of the Coronaviridae family, which is responsible for the COVID-19 pandemic followed by unprecedented global societal and economic disruptive impact. The innate immune system is the body's first line of defense against invading pathogens and is induced by a variety of cellular receptors that sense viral components. However, various strategies are exploited by SARS-CoV-2 to disrupt the antiviral innate immune responses. Innate immune dysfunction is characterized by the weak generation of type I interferons (IFNs) and the hypersecretion of pro-inflammatory cytokines, leading to mortality and organ injury in patients with COVID-19. This review summarizes the existing understanding of the mutual effects between SARS-CoV-2 and the type I IFN (IFN-α/β) responses, emphasizing the relationship between host innate immune signaling and viral proteases with an insight on tackling potential therapeutic targets.
Keywords: SARS-CoV-2; innate immune response; type I interferons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

