The Expression Level of HIV-1 Vif Is Optimized by Nucleotide Changes in the Genomic SA1D2prox Region during the Viral Adaptation Process
- PMID: 34696508
- PMCID: PMC8537775
- DOI: 10.3390/v13102079
The Expression Level of HIV-1 Vif Is Optimized by Nucleotide Changes in the Genomic SA1D2prox Region during the Viral Adaptation Process
Abstract
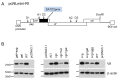
HIV-1 Vif plays an essential role in viral replication by antagonizing anti-viral cellular restriction factors, a family of APOBEC3 proteins. We have previously shown that naturally-occurring single-nucleotide mutations in the SA1D2prox region, which surrounds the splicing acceptor 1 and splicing donor 2 sites of the HIV-1 genome, dramatically alter the Vif expression level, resulting in variants with low or excessive Vif expression. In this study, we investigated how these HIV-1 variants with poor replication ability adapt and evolve under the pressure of APOBEC3 proteins. Adapted clones obtained through adaptation experiments exhibited an altered replication ability and Vif expression level compared to each parental clone. While various mutations were present throughout the viral genome, all replication-competent adapted clones with altered Vif expression levels were found to bear them within SA1D2prox, without exception. Indeed, the mutations identified within SA1D2prox were responsible for changes in the Vif expression levels and altered the splicing pattern. Moreover, for samples collected from HIV-1-infected patients, we showed that the nucleotide sequences of SA1D2prox can be chronologically changed and concomitantly affect the Vif expression levels. Taken together, these results demonstrated the importance of the SA1D2prox nucleotide sequence for modulating the Vif expression level during HIV-1 replication and adaptation.
Keywords: HIV-1; SA1D2prox; Vif expression; adaptation; nucleotide sequence; splicing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



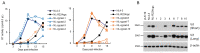






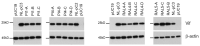
Similar articles
-
Natural Single-Nucleotide Variations in the HIV-1 Genomic SA1prox Region Can Alter Viral Replication Ability by Regulating Vif Expression Levels.J Virol. 2016 Apr 14;90(9):4563-4578. doi: 10.1128/JVI.02939-15. Print 2016 May. J Virol. 2016. PMID: 26912631 Free PMC article.
-
Expression Level of HIV-1 Vif Can Be Fluctuated by Natural Nucleotide Variations in the vif-Coding and Regulatory SA1D2prox Sequences of the Proviral Genome.Front Microbiol. 2019 Nov 28;10:2758. doi: 10.3389/fmicb.2019.02758. eCollection 2019. Front Microbiol. 2019. PMID: 31849897 Free PMC article.
-
Production of HIV-1 vif mRNA Is Modulated by Natural Nucleotide Variations and SLSA1 RNA Structure in SA1D2prox Genomic Region.Front Microbiol. 2017 Dec 18;8:2542. doi: 10.3389/fmicb.2017.02542. eCollection 2017. Front Microbiol. 2017. PMID: 29326677 Free PMC article.
-
Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all.J Mol Biol. 2014 Mar 20;426(6):1220-45. doi: 10.1016/j.jmb.2013.10.033. Epub 2013 Nov 2. J Mol Biol. 2014. PMID: 24189052 Free PMC article. Review.
-
Running loose or getting lost: how HIV-1 counters and capitalizes on APOBEC3-induced mutagenesis through its Vif protein.Viruses. 2012 Nov 14;4(11):3132-61. doi: 10.3390/v4113132. Viruses. 2012. PMID: 23202519 Free PMC article. Review.
Cited by
-
Altered HIV-1 mRNA Splicing Due to Drug-Resistance-Associated Mutations in Exon 2/2b.Int J Mol Sci. 2021 Dec 23;23(1):156. doi: 10.3390/ijms23010156. Int J Mol Sci. 2021. PMID: 35008581 Free PMC article.
-
Involvement of a Rarely Used Splicing SD2b Site in the Regulation of HIV-1 vif mRNA Production as Revealed by a Growth-Adaptive Mutation.Viruses. 2023 Dec 14;15(12):2424. doi: 10.3390/v15122424. Viruses. 2023. PMID: 38140666 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

