Molecular Characterization and Taxonomic Assignment of Three Phage Isolates from a Collection Infecting Pseudomonas syringae pv. actinidiae and P. syringae pv. phaseolicola from Northern Italy
- PMID: 34696512
- PMCID: PMC8537276
- DOI: 10.3390/v13102083
Molecular Characterization and Taxonomic Assignment of Three Phage Isolates from a Collection Infecting Pseudomonas syringae pv. actinidiae and P. syringae pv. phaseolicola from Northern Italy
Abstract
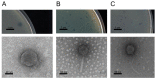
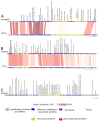
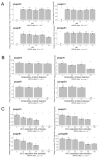
Bacterial kiwifruit vine disease (Pseudomonas syringae pv. actinidiae, Psa) and halo blight of bean (P. syringae pv. phaseolicola, Pph) are routinely treated with copper, leading to environmental pollution and bacterial copper resistance. An alternative sustainable control method could be based on bacteriophages, as phage biocontrol offers high specificity and does not result in the spread of toxic residues into the environment or the food chain. In this research, specific phages suitable for phage-based biocontrol strategies effective against Psa and Pph were isolated and characterized. In total, sixteen lytic Pph phage isolates and seven lytic Psa phage isolates were isolated from soil in Piedmont and Veneto in northern Italy. Genome characterization of fifteen selected phages revealed that the isolated Pph phages were highly similar and could be considered as isolates of a novel species, whereas the isolated Psa phages grouped into four distinct clades, two of which represent putative novel species. No lysogeny-, virulence- or toxin-related genes were found in four phages, making them suitable for potential biocontrol purposes. A partial biological characterization including a host range analysis was performed on a representative subset of these isolates. This analysis was a prerequisite to assess their efficacy in greenhouse and in field trials, using different delivery strategies.
Keywords: Pph; Psa; Pseudomonas syringae; bean; bean halo blight; biocontrol; kiwifruit; kiwifruit canker; phage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Identification of bacteriophages for biocontrol of the kiwifruit canker phytopathogen Pseudomonas syringae pv. actinidiae.Appl Environ Microbiol. 2014 Apr;80(7):2216-28. doi: 10.1128/AEM.00062-14. Epub 2014 Jan 31. Appl Environ Microbiol. 2014. PMID: 24487530 Free PMC article.
-
Isolation and Characterization of Bacteriophages Against Pseudomonas syringae pv. actinidiae Causing Bacterial Canker Disease in Kiwifruit.J Microbiol Biotechnol. 2016 Feb;26(2):385-93. doi: 10.4014/jmb.1509.09012. J Microbiol Biotechnol. 2016. PMID: 26628254
-
Isolation and partial characterization of bacteriophages infecting Pseudomonas syringae pv. actinidiae, causal agent of kiwifruit bacterial canker.J Basic Microbiol. 2014 Nov;54(11):1210-21. doi: 10.1002/jobm.201300951. Epub 2014 May 9. J Basic Microbiol. 2014. PMID: 24810619
-
Advancements in the Use of Bacteriophages to Combat the Kiwifruit Canker Phytopathogen Pseudomonas syringae pv. actinidiae.Viruses. 2022 Dec 2;14(12):2704. doi: 10.3390/v14122704. Viruses. 2022. PMID: 36560706 Free PMC article. Review.
-
Kiwifruit bacterial canker: an integrative view focused on biocontrol strategies.Planta. 2021 Jan 27;253(2):49. doi: 10.1007/s00425-020-03549-1. Planta. 2021. PMID: 33502587 Review.
Cited by
-
Back to the Roots: Agrobacterium-Specific Phages Show Potential to Disinfect Nutrient Solution from Hydroponic Greenhouses.Appl Environ Microbiol. 2023 Apr 26;89(4):e0021523. doi: 10.1128/aem.00215-23. Epub 2023 Apr 3. Appl Environ Microbiol. 2023. PMID: 37010433 Free PMC article.
-
Isolation, characterization and genome analysis of an orphan phage FoX4 of the new Foxquatrovirus genus.BMC Microbiol. 2022 Dec 13;22(1):304. doi: 10.1186/s12866-022-02719-3. BMC Microbiol. 2022. PMID: 36513996 Free PMC article.
-
Lytic Spectra of Tailed Bacteriophages: A Systematic Review and Meta-Analysis.Viruses. 2024 Dec 4;16(12):1879. doi: 10.3390/v16121879. Viruses. 2024. PMID: 39772189 Free PMC article.
-
Biological and Molecular Characterization of the Lytic Bacteriophage SoKa against Pseudomonas syringae pv. syringae, Causal Agent of Citrus Blast and Black Pit in Tunisia.Viruses. 2022 Sep 2;14(9):1949. doi: 10.3390/v14091949. Viruses. 2022. PMID: 36146756 Free PMC article.
-
Detection, Diagnosis, and Preventive Management of the Bacterial Plant Pathogen Pseudomonas syringae.Plants (Basel). 2023 Apr 25;12(9):1765. doi: 10.3390/plants12091765. Plants (Basel). 2023. PMID: 37176823 Free PMC article. Review.
References
-
- Froud K.J., Everett K.R., Tyson J.L., Beresford R.M., Cogger N. Review of the risk factors associated with kiwifruit bacterial canker caused by Pseudomonas syringae pv actinidiae. N. Z. Plant Prot. 2015;68:313–327. doi: 10.30843/nzpp.2015.68.5828. - DOI
-
- Taylor J.D., Teverson D.M., Davis J.H.C. Sources of resistance to Pseudomonas syringae pv phaseolicola races in Phaseolus vulgaris. Plant Pathol. 1996;45:479–485. doi: 10.1046/j.1365-3059.1996.d01-148.x. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

