Comparison of PCR versus PCR-Free DNA Library Preparation for Characterising the Human Faecal Virome
- PMID: 34696523
- PMCID: PMC8537689
- DOI: 10.3390/v13102093
Comparison of PCR versus PCR-Free DNA Library Preparation for Characterising the Human Faecal Virome
Abstract
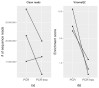
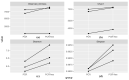
The human intestinal microbiota is abundant in viruses, comprising mainly bacteriophages, occasionally outnumbering bacteria 10:1 and is termed the virome. Due to their high genetic diversity and the lack of suitable tools and reference databases, the virome remains poorly characterised and is often referred to as "viral dark matter". However, the choice of sequencing platforms, read lengths and library preparation make study design challenging with respect to the virome. Here we have compared the use of PCR and PCR-free methods for sequence-library construction on the Illumina sequencing platform for characterising the human faecal virome. Viral DNA was extracted from faecal samples of three healthy donors and sequenced. Our analysis shows that most variation was reflecting the individually specific faecal virome. However, we observed differences between PCR and PCR-free library preparation that affected the recovery of low-abundance viral genomes. Using three faecal samples in this study, the PCR library preparation samples led to a loss of lower-abundance vOTUs evident in their PCR-free pairs (vOTUs 128, 6202 and 8364) and decreased the alpha-diversity indices (Chao1 p-value = 0.045 and Simpson p-value = 0.044). Thus, differences between PCR and PCR-free methods are important to consider when investigating "rare" members of the gut virome, with these biases likely negligible when investigating moderately and highly abundant viruses.
Keywords: PCR bias; bacteriophage; virome.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


Similar articles
-
Nanopore and Illumina sequencing reveal different viral populations from human gut samples.Microb Genom. 2024 Apr;10(4):001236. doi: 10.1099/mgen.0.001236. Microb Genom. 2024. PMID: 38683195 Free PMC article.
-
A single-stranded based library preparation method for virome characterization.Microbiome. 2024 Oct 24;12(1):219. doi: 10.1186/s40168-024-01935-5. Microbiome. 2024. PMID: 39449043 Free PMC article.
-
Complementary insights into gut viral genomes: a comparative benchmark of short- and long-read metagenomes using diverse assemblers and binners.Microbiome. 2024 Dec 20;12(1):260. doi: 10.1186/s40168-024-01981-z. Microbiome. 2024. PMID: 39707560 Free PMC article.
-
The Human Virome: Viral Metagenomics, Relations with Human Diseases, and Therapeutic Applications.Viruses. 2022 Jan 28;14(2):278. doi: 10.3390/v14020278. Viruses. 2022. PMID: 35215871 Free PMC article. Review.
-
The human virome: assembly, composition and host interactions.Nat Rev Microbiol. 2021 Aug;19(8):514-527. doi: 10.1038/s41579-021-00536-5. Epub 2021 Mar 30. Nat Rev Microbiol. 2021. PMID: 33785903 Free PMC article. Review.
Cited by
-
Challenges and insights in the exploration of the low abundance human ocular surface microbiome.Front Cell Infect Microbiol. 2023 Sep 1;13:1232147. doi: 10.3389/fcimb.2023.1232147. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37727808 Free PMC article.
-
Nanopore and Illumina sequencing reveal different viral populations from human gut samples.Microb Genom. 2024 Apr;10(4):001236. doi: 10.1099/mgen.0.001236. Microb Genom. 2024. PMID: 38683195 Free PMC article.
-
Assessing Bias and Reproducibility of Viral Metagenomics Methods for the Combined Detection of Faecal RNA and DNA Viruses.Viruses. 2025 Jan 23;17(2):155. doi: 10.3390/v17020155. Viruses. 2025. PMID: 40006910 Free PMC article.
-
An Exploration of the Relationship Between Gut Virome and Cardiovascular Disease: A Comprehensive Review.Rev Cardiovasc Med. 2025 Jun 24;26(6):36386. doi: 10.31083/RCM36386. eCollection 2025 Jun. Rev Cardiovasc Med. 2025. PMID: 40630433 Free PMC article. Review.
-
Clinical application of amplification-based versus amplification-free metagenomic next-generation sequencing test in infectious diseases.Front Cell Infect Microbiol. 2023 Nov 29;13:1138174. doi: 10.3389/fcimb.2023.1138174. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38094744 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- BBS/E/F/000PR10353/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10355/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10356 410/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10356/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/R012490/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

