CircCD44 plays oncogenic roles in triple-negative breast cancer by modulating the miR-502-5p/KRAS and IGF2BP2/Myc axes
- PMID: 34696797
- PMCID: PMC8543802
- DOI: 10.1186/s12943-021-01444-1
CircCD44 plays oncogenic roles in triple-negative breast cancer by modulating the miR-502-5p/KRAS and IGF2BP2/Myc axes
Erratum in
-
Correction: CircCD44 plays oncogenic roles in triple-negative breast cancer by modulating the miR-502-5p/KRAS and IGF2BP2/Myc axes.Mol Cancer. 2025 Feb 14;24(1):46. doi: 10.1186/s12943-025-02257-2. Mol Cancer. 2025. PMID: 39953598 Free PMC article. No abstract available.
Abstract
Background: Emerging studies have revealed the potent functions of circRNAs in breast cancer tumorigenesis. However, the biogenesis, biofunction and mechanism of circRNAs in triple-negative breast cancer (TNBC) are largely unknown.
Methods: High-throughput RNA sequencing was applied to identify dysregulated circRNAs in TNBCs and paired normal tissues. RNA pulldown and luciferase assays were performed to investigate the interaction between circular CD44 (circCD44, also annotated as hsa_circ_0021735) and miR-502-5p. RNA pulldown and RIP assays were used to investigate the interaction between circCD44 and IGF2BP2. Cell viability, colony formation, migration/invasion assays and in vivo tumorigenesis were used to investigate circCD44 biological functions.
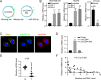
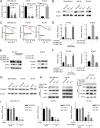
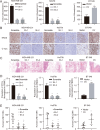
Results: CircCD44 is an uncharacterized circRNA, which is highly expressed in TNBC, and its expression is negatively correlated with the prognosis of TNBC patients. CircCD44 promotes TNBC proliferation, migration, invasion and tumorigenesis at least partially by sponging miR-502-5p and interacting with IGF2BP2.
Conclusion: Our data suggested that overexpressed circCD44 promotes TNBC progression. CircCD44 is potentially a novel diagnostic and therapeutic marker for TNBC patients.
Keywords: IGF2BP2; KRAS; MYC; TNBC; circCD44; circRNAs.
© 2021. The Author(s).
Conflict of interest statement
There are no potential conflicts of interest to disclose.
Figures





References
-
- Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. - PubMed
-
- Masuda N, et al. Adjuvant Capecitabine for breast Cancer after preoperative chemotherapy. N Engl J Med. 2017;376(22):2147–59. - PubMed
-
- Mougalian SS, et al. Use of neoadjuvant chemotherapy for patients with stage I to III breast cancer in the United States. Cancer. 2015;121(15):2544–52. - PubMed
-
- Pascual J, Turner NC. Targeting the PI3-kinase pathway in triple-negative breast cancer. Ann Oncol. 2019;30(7):1051–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

