Long Noncoding RNA ZFAS1 Promotes Progression of Oral Squamous Cell Carcinoma Through Targeting miR-6499-3p/CCL5 Axis
- PMID: 34697152
- PMCID: PMC8627759
- DOI: 10.21873/invivo.12616
Long Noncoding RNA ZFAS1 Promotes Progression of Oral Squamous Cell Carcinoma Through Targeting miR-6499-3p/CCL5 Axis
Abstract
Background: Oral squamous cell carcinoma (OSCC) ranks sixth among malignancies in the world, and there are 200,000 new cases annually, rendering OSCC a significant global public health issue that has caused great burdens on patients, society, and the economy. Despite great progress in diagnosis and treatment methods, patient survival has not been greatly enhanced. Hence, there is an urgent need to identify novel targets that can serve as early diagnostic and therapeutic biomarkers for OSCC. Long non-coding RNAs (lncRNAs) participate in several cancer types, including OSCC. This work identified the competing endogenous RNA network related to lncRNA zinc finger nuclear transcription factor, X-box binding 1-type containing 1 antisense RNA 1 (ZFAS1) in OSCC, as well as the corresponding downstream targets.
Materials and methods: Firstly, we identified the lncRNA ZFAS1 levels in OSCC cells and tissues and confirmed its relationship to tumor progression. Secondly, we identified a lncRNA-miRNA-mRNA network, which was closely associated with OSCC development using bioinformatics methods. Next, our hypothesis that lncRNA ZFAS1 modulates OSCC progression was verified with in vitro and in vivo experiments.
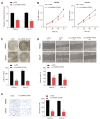
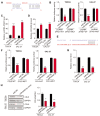
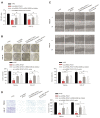
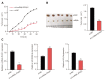
Results: Firstly, we found lncRNA ZFAS1 expression increased within OSCC cells and tissues and was positively associated with tumor progression. Secondly, its lncRNA-miRNA-mRNA network was determined, and the target of ZFAS1 was identified as miR-6499-3p/C-C motif chemokine ligand 5 (CCL5). Mechanistically, we found that ZFAS1 up-regulated CCL5 by competitively sponging miR-6499-3p. Further studies demonstrated that ZFAS1 promoted tumor progression in vivo and in vitro.
Conclusion: Our results indicate that ZFAS1 serves as a crucial oncogenic factor in OSCC occurrence and development and may therefore serve as a possible therapeutic target for OSCC.
Keywords: CCL5; Oral squamous cell carcinoma; ZFAS1; ceRNA; lncRNA.
Copyright © 2021 International Institute of Anticancer Research (Dr. George J. Delinasios), All rights reserved.
Conflict of interest statement
The Authors declare no competing financial interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
