Sex-specific genetic regulation of adipose mitochondria and metabolic syndrome by Ndufv2
- PMID: 34697471
- PMCID: PMC8909918
- DOI: 10.1038/s42255-021-00481-w
Sex-specific genetic regulation of adipose mitochondria and metabolic syndrome by Ndufv2
Abstract
We have previously suggested a central role for mitochondria in the observed sex differences in metabolic traits. However, the mechanisms by which sex differences affect adipose mitochondrial function and metabolic syndrome are unclear. Here we show that in both mice and humans, adipose mitochondrial functions are elevated in females and are strongly associated with adiposity, insulin resistance and plasma lipids. Using a panel of diverse inbred strains of mice, we identify a genetic locus on mouse chromosome 17 that controls mitochondrial mass and function in adipose tissue in a sex- and tissue-specific manner. This locus contains Ndufv2 and regulates the expression of at least 89 mitochondrial genes in females, including oxidative phosphorylation genes and those related to mitochondrial DNA content. Overexpression studies indicate that Ndufv2 mediates these effects by regulating supercomplex assembly and elevating mitochondrial reactive oxygen species production, which generates a signal that increases mitochondrial biogenesis.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures

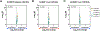
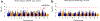




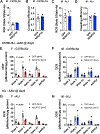







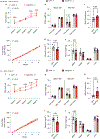

References
Publication types
MeSH terms
Substances
Grants and funding
- K99 DK120875/DK/NIDDK NIH HHS/United States
- R01 HL125863/HL/NHLBI NIH HHS/United States
- R01 HL147187/HL/NHLBI NIH HHS/United States
- R01 HL144651/HL/NHLBI NIH HHS/United States
- R01 DK117850/DK/NIDDK NIH HHS/United States
- P01 HL028481/HL/NHLBI NIH HHS/United States
- U54 DK120342/DK/NIDDK NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- R01 AA026914/AA/NIAAA NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- P30 DK041301/DK/NIDDK NIH HHS/United States
- R00 DK120875/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

