Unlocking SARS-CoV-2 detection in low- and middle-income countries
- PMID: 34697612
- PMCID: PMC8529268
- DOI: 10.1016/j.crmeth.2021.100093
Unlocking SARS-CoV-2 detection in low- and middle-income countries
Abstract
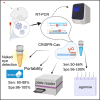
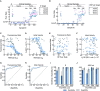
Low- and middle-income countries (LMICs) are significantly affected by SARS-CoV-2, partially due to their limited capacity for local production and implementation of molecular testing. Here, we provide detailed methods and validation of a molecular toolkit that can be readily produced and deployed using laboratory equipment available in LMICs. Our results show that lab-scale production of enzymes and nucleic acids can supply over 50,000 tests per production batch. The optimized one-step RT-PCR coupled to CRISPR-Cas12a-mediated detection showed a limit of detection of 102 ge/μL in a turnaround time of 2 h. The clinical validation indicated an overall sensitivity of 80%-88%, while for middle and high viral load samples (Cq ≤ 31) the sensitivity was 92%-100%. The specificity was 96%-100% regardless of viral load. Furthermore, we show that the toolkit can be used with the mobile laboratory Bento Lab, potentially enabling LMICs to implement detection services in unattended remote regions.
Keywords: COVID-19; CRISPR; LMICs; RT-PCR; SARS-CoV-2; fluorescence; molecular testing.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures




Comment in
-
Open access methods and protocols promote open science in a pandemic.STAR Protoc. 2022 Mar 2;3(1):101226. doi: 10.1016/j.xpro.2022.101226. eCollection 2022 Mar 18. STAR Protoc. 2022. PMID: 35284832 Free PMC article.
References
-
- Abbott S., Hellewell J., Thompson R.N., Sherratt K., Gibbs H.P., Bosse N.I., Munday J.D., Meakin S., Doughty E.L., Chun J.Y., et al. Estimating the time-varying reproduction number of SARS-CoV-2 using national and subnational case counts. Wellcome Open Res. 2020;5:112.
-
- Aslanzadeh J. Preventing PCR amplification carryover contamination in a clinical laboratory. Ann. Clin. Lab. Sci. 2004;34:389–396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

