Novel Selection Approaches to Identify Antibodies Targeting Neoepitopes on the C5b6 Intermediate Complex to Inhibit Membrane Attack Complex Formation
- PMID: 34698051
- PMCID: PMC8544208
- DOI: 10.3390/antib10040039
Novel Selection Approaches to Identify Antibodies Targeting Neoepitopes on the C5b6 Intermediate Complex to Inhibit Membrane Attack Complex Formation
Abstract
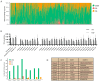
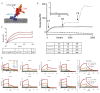
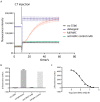
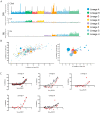
The terminal pathway of complement is implicated in the pathology of multiple diseases and its inhibition is, therefore, an attractive therapeutic proposition. The practicalities of inhibiting this pathway, however, are challenging, as highlighted by the very few molecules in the clinic. The proteins are highly abundant, and assembly is mediated by high-affinity protein-protein interactions. One strategy is to target neoepitopes that are present transiently and only exist on active or intermediate complexes but not on the abundant native proteins. Here, we describe an antibody discovery campaign that generated neoepitope-specific mAbs against the C5b6 complex, a stable intermediate complex in terminal complement complex assembly. We used a highly diverse yeast-based antibody library of fully human IgGs to screen against soluble C5b6 antigen and successfully identified C5b6 neoepitope-specific antibodies. These antibodies were diverse, showed good binding to C5b6, and inhibited membrane attack complex (MAC) formation in a solution-based assay. However, when tested in a more physiologically relevant membrane-based assay these antibodies failed to inhibit MAC formation. Our data highlight the feasibility of identifying neoepitope binding mAbs, but also the technical challenges associated with the identification of functionally relevant, neoepitope-specific inhibitors of the terminal pathway.
Keywords: antibody discovery; complement; neoepitope; terminal pathway; therapeutic antibody.
Conflict of interest statement
E.-M.N., M.F., S.J.K, T.D.B., L.S., E.K.H.D., R.P.B., D.A.G., K.A.W., N.T., A.S., J.L.B. and J.E.C. are employees and shareholders of GSK plc. J.P.H., A.T. and C.L.H. are now employees of Gyroscope Therapeutics, AstraZeneca and Newcastle University (Translational and Clinical Research Institute), respectively.
Figures


Similar articles
-
Interaction between complement proteins C5b-7 and erythrocyte membrane sialic acid.J Exp Med. 1996 Oct 1;184(4):1225-32. doi: 10.1084/jem.184.4.1225. J Exp Med. 1996. PMID: 8879193 Free PMC article.
-
Hydrolysis of myelin basic protein in human myelin by terminal complement complexes.J Biol Chem. 1988 May 25;263(15):7228-34. J Biol Chem. 1988. PMID: 2452821
-
Serum amyloid P component forms a stable complex with human C5b6.J Immunol. 1997 Apr 15;158(8):3830-5. J Immunol. 1997. PMID: 9103450
-
Use of anti-neoepitope antibodies for the analysis of degradative events in cartilage and the molecular basis for neoepitope specificity.Biochem Soc Symp. 2003;(70):107-14. doi: 10.1042/bss0700107. Biochem Soc Symp. 2003. PMID: 14587286 Review.
-
Inhibiting the C5-C5a receptor axis.Mol Immunol. 2011 Aug;48(14):1631-42. doi: 10.1016/j.molimm.2011.04.014. Epub 2011 May 6. Mol Immunol. 2011. PMID: 21549429 Review.
Cited by
-
Action of the Terminal Complement Pathway on Cell Membranes.J Membr Biol. 2025 Aug;258(4):269-304. doi: 10.1007/s00232-025-00343-6. Epub 2025 Mar 23. J Membr Biol. 2025. PMID: 40122920 Free PMC article. Review.
References
-
- Taylor P.R., Carugati A., Fadok V.A., Cook H.T., Andrews M., Carroll M.C., Savill J.S., Henson P.M., Botto M., Walport M.J. A hierarchical role for classical pathway complement proteins in the clearance of apoptotic cells in vivo. J. Exp. Med. 2000;192:359–366. doi: 10.1084/jem.192.3.359. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

