Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant
- PMID: 34698504
- PMCID: PMC10763652
- DOI: 10.1126/science.abl9463
Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant
Abstract
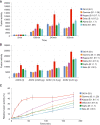
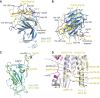
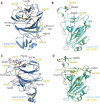
The Delta variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has outcompeted previously prevalent variants and become a dominant strain worldwide. We report the structure, function, and antigenicity of its full-length spike (S) trimer as well as those of the Gamma and Kappa variants, and compare their characteristics with the G614, Alpha, and Beta variants. Delta S can fuse membranes more efficiently at low levels of cellular receptor angiotensin converting enzyme 2 (ACE2), and its pseudotyped viruses infect target cells substantially faster than the other five variants, possibly accounting for its heightened transmissibility. Each variant shows different rearrangement of the antigenic surface of the amino-terminal domain of the S protein but only makes produces changes in the receptor binding domain (RBD), making the RBD a better target for therapeutic antibodies.
Figures


Update of
-
Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.bioRxiv [Preprint]. 2021 Aug 17:2021.08.17.456689. doi: 10.1101/2021.08.17.456689. bioRxiv. 2021. Update in: Science. 2021 Dec 10;374(6573):1353-1360. doi: 10.1126/science.abl9463. PMID: 34426810 Free PMC article. Updated. Preprint.
Similar articles
-
Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.Science. 2021 Aug 6;373(6555):642-648. doi: 10.1126/science.abi9745. Epub 2021 Jun 24. Science. 2021. PMID: 34168070 Free PMC article.
-
Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants.Science. 2021 Dec 24;374(6575):1621-1626. doi: 10.1126/science.abl8506. Epub 2021 Nov 9. Science. 2021. PMID: 34751595 Free PMC article.
-
SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.Science. 2022 Feb 18;375(6582):760-764. doi: 10.1126/science.abn7760. Epub 2022 Jan 20. Science. 2022. PMID: 35050643 Free PMC article.
-
Structural Evaluation of the Spike Glycoprotein Variants on SARS-CoV-2 Transmission and Immune Evasion.Int J Mol Sci. 2021 Jul 10;22(14):7425. doi: 10.3390/ijms22147425. Int J Mol Sci. 2021. PMID: 34299045 Free PMC article. Review.
-
Targeting SARS-CoV2 Spike Protein Receptor Binding Domain by Therapeutic Antibodies.Biomed Pharmacother. 2020 Oct;130:110559. doi: 10.1016/j.biopha.2020.110559. Epub 2020 Aug 1. Biomed Pharmacother. 2020. PMID: 32768882 Free PMC article. Review.
Cited by
-
Differential proinflammatory activities of Spike proteins of SARS-CoV-2 variants of concern.Sci Adv. 2022 Sep 16;8(37):eabo0732. doi: 10.1126/sciadv.abo0732. Epub 2022 Sep 16. Sci Adv. 2022. PMID: 36112681 Free PMC article.
-
Development of highly effective LCB1-based lipopeptides targeting the spike receptor-binding motif of SARS-CoV-2.Antiviral Res. 2023 Mar;211:105541. doi: 10.1016/j.antiviral.2023.105541. Epub 2023 Jan 20. Antiviral Res. 2023. PMID: 36682464 Free PMC article.
-
Imaging Severity COVID-19 Assessment in Vaccinated and Unvaccinated Patients: Comparison of the Different Variants in a High Volume Italian Reference Center.J Pers Med. 2022 Jun 10;12(6):955. doi: 10.3390/jpm12060955. J Pers Med. 2022. PMID: 35743740 Free PMC article.
-
Breakthrough COVID-19 Infections in the US: Implications for Prolonging the Pandemic.Vaccines (Basel). 2022 May 11;10(5):755. doi: 10.3390/vaccines10050755. Vaccines (Basel). 2022. PMID: 35632512 Free PMC article. Review.
-
Inhibitors of Activin Receptor-like Kinase 5 Interfere with SARS-CoV-2 S-Protein Processing and Spike-Mediated Cell Fusion via Attenuation of Furin Expression.Viruses. 2022 Jun 15;14(6):1308. doi: 10.3390/v14061308. Viruses. 2022. PMID: 35746781 Free PMC article.
References
-
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). 10.1038/s41586-020-2012-7 - DOI - PMC - PubMed
-
- Cai Y., Zhang J., Xiao T., Lavine C. L., Rawson S., Peng H., Zhu H., Anand K., Tong P., Gautam A., Lu S., Sterling S. M., Walsh R. M. Jr., Rits-Volloch S., Lu J., Wesemann D. R., Yang W., Seaman M. S., Chen B., Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants. Science 373, 642–648 (2021). 10.1126/science.abi9745 - DOI - PMC - PubMed
-
- Mlcochova P., Kemp S., Dhar M. S., Papa G., Meng B., Mishra S., Whittaker C., Mellan T., Ferreira I., Datir R., Collier D. A., Singh S., Pandey R., Marwal R., Datta M., Sengupta S., Ponnusamy K., Radhakrishnan V. S., Abdullahi A., Goonawardne N., Brown J., Charles O., Chattopadhyay P., Devi P., Caputo D., Peacock T., Wattal C., Goel N., Vaishya R., Agarwal M., The Indian SARS-CoV-2 Genomics Consortium (INSACOG), CITIID-NIHR BioResource COVID-19 Collaboration, A. Mavousian, H. Lee, W. S. Barcla, S. Bhatt, S. Flaxman, L. James, P. Rakshit, A. Agrawal, R. K. Gupta, SARS-CoV-2 B.1.617.2 Delta variant replication, sensitivity to neutralising antibodies and vaccine breakthrough. Nature 599, 114–119 [Preprint] (2021). 10.21203/rs.3.rs-637724/v1 - DOI - PubMed
-
- Earnest R., Uddin R., Matluk N., Renzette N., Siddle K. J., Loreth C., Adams G., Tomkins-Tinch C. H., Petrone M. E., Rothman J. E., Breban M. I., Koch R. T., Billig K., Fauver J. R., Vogels C. B. F., Turbett S., Bilguvar K., De Kumar B., Landry M. L., Peaper D. R., Kelly K., Omerza G., Grieser H., Meak S., Martha J., Dewey H. H., Kales S., Berenzy D., Carpenter-Azevedo K., King E., Huard R. C., Smole S. C., Brown C. M., Fink T., Lang A. S., Gallagher G. R., Sabeti P. C., Gabriel S., MacInnis B. L., Tewhey R., Adams M. D., Park D. J., Lemieux J. E., Grubaugh N. D., Comparative transmissibility of SARS-CoV-2 variants Delta and Alpha in New England, USA. medRxiv, 2021.10.06.21264641 [Preprint] (2021). 10.1101/2021.10.06.21264641 - DOI - PMC - PubMed
-
- Dagpunar J., Interim estimates of increased transmissibility, growth rate, and reproduction number of the Covid-19 B.1.617.2 variant of concern in the United Kingdom. medRxiv, 2021.2006.2003.21258293 [Preprint] (2021). 10.1101/2021.06.03.21258293 - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

