Progress towards completing the mutant mouse null resource
- PMID: 34698892
- PMCID: PMC8913489
- DOI: 10.1007/s00335-021-09905-0
Progress towards completing the mutant mouse null resource
Abstract
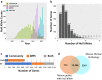
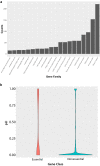
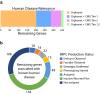
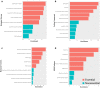
The generation of a comprehensive catalog of null alleles covering all protein-coding genes is the goal of the International Mouse Phenotyping Consortium. Over the past 20 years, significant progress has been made towards achieving this goal through the combined efforts of many large-scale programs that built an embryonic stem cell resource to generate knockout mice and more recently employed CRISPR/Cas9-based mutagenesis to delete critical regions predicted to result in frameshift mutations, thus, ablating gene function. The IMPC initiative builds on prior and ongoing work by individual research groups creating gene knockouts in the mouse. Here, we analyze the collective efforts focusing on the combined null allele resource resulting from strains developed by the research community and large-scale production programs. Based upon this pooled analysis, we examine the remaining fraction of protein-coding genes focusing on clearly defined mouse-human orthologs as the highest priority for completing the mutant mouse null resource. In summary, we find that there are less than 3400 mouse-human orthologs remaining in the genome without a targeted null allele that can be further prioritized to achieve our overall goal of the complete functional annotation of the protein-coding portion of a mammalian genome.
© 2021. The Author(s).
Conflict of interest statement
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Figures

Similar articles
-
Comparative analysis of single-stranded DNA donors to generate conditional null mouse alleles.BMC Biol. 2018 Jun 21;16(1):69. doi: 10.1186/s12915-018-0529-0. BMC Biol. 2018. PMID: 29925370 Free PMC article.
-
Impact of essential genes on the success of genome editing experiments generating 3313 new genetically engineered mouse lines.Sci Rep. 2024 Sep 30;14(1):22626. doi: 10.1038/s41598-024-72418-8. Sci Rep. 2024. PMID: 39349521 Free PMC article.
-
Beyond knockouts: the International Knockout Mouse Consortium delivers modular and evolving tools for investigating mammalian genes.Mamm Genome. 2015 Oct;26(9-10):456-66. doi: 10.1007/s00335-015-9598-3. Epub 2015 Sep 4. Mamm Genome. 2015. PMID: 26340938
-
The mouse resource at National Resource Center for Mutant Mice.Mamm Genome. 2022 Mar;33(1):143-156. doi: 10.1007/s00335-021-09940-x. Epub 2022 Feb 9. Mamm Genome. 2022. PMID: 35138443 Review.
-
Predicting human disease mutations and identifying drug targets from mouse gene knockout phenotyping campaigns.Dis Model Mech. 2019 May 7;12(5):dmm038224. doi: 10.1242/dmm.038224. Dis Model Mech. 2019. PMID: 31064765 Free PMC article. Review.
Cited by
-
X-linked palindromic gene families 4930567H17Rik and Mageb5 are dispensable for male mouse fertility.Sci Rep. 2022 May 20;12(1):8554. doi: 10.1038/s41598-022-12433-9. Sci Rep. 2022. PMID: 35595785 Free PMC article.
-
Essential genes: a cross-species perspective.Mamm Genome. 2023 Sep;34(3):357-363. doi: 10.1007/s00335-023-09984-1. Epub 2023 Mar 10. Mamm Genome. 2023. PMID: 36897351 Free PMC article. Review.
-
Contribution of model organism phenotypes to the computational identification of human disease genes.Dis Model Mech. 2022 Jul 1;15(7):dmm049441. doi: 10.1242/dmm.049441. Epub 2022 Aug 3. Dis Model Mech. 2022. PMID: 35758016 Free PMC article.
-
De novo PAM generation to reach initially inaccessible target sites for base editing.Development. 2023 Jan 15;150(2):dev201115. doi: 10.1242/dev.201115. Epub 2023 Jan 23. Development. 2023. PMID: 36683434 Free PMC article.
-
A graph theoretical approach to experimental prioritization in genome-scale investigations.Mamm Genome. 2024 Dec;35(4):724-733. doi: 10.1007/s00335-024-10066-z. Epub 2024 Aug 27. Mamm Genome. 2024. PMID: 39191873 Free PMC article.
References
-
- Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, Grieder FB, Heintz N, Hicks G, Insel TR, Joyner A, Koller BH, Lloyd KC, Magnuson T, Moore MW, Nagy A, Pollock JD, Roses AD, Sands AT, Seed B, Skarnes WC, Snoddy J, Soriano P, Stewart DJ, Stewart F, Stillman B, Varmus H, Varticovski L, Verma IM, Vogt TF, von Melchner H, Witkowski J, Woychik RP, Wurst W, Yancopoulos GD, Young SG, Zambrowicz B. The knockout mouse project. Nat Genet. 2004;36:921–924. - PMC - PubMed
-
- Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, Barbacid M, Berns A, Bradley A, Brown S, Carmeliet P, Chambon P, Cox R, Davidson D, Davies K, Duboule D, Forejt J, Granucci F, Hastie N, de Angelis MH, Jackson I, Kioussis D, Kollias G, Lathrop M, Lendahl U, Malumbres M, von Melchner H, Muller W, Partanen J, Ricciardi-Castagnoli P, Rigby P, Rosen B, Rosenthal N, Skarnes B, Stewart AF, Thornton J, Tocchini-Valentini G, Wagner E, Wahli W, Wurst W. The European dimension for the mouse genome mutagenesis program. Nat Genet. 2004;36:925–927. - PMC - PubMed
-
- Bartha I, di Iulio J, Venter JC, Telenti A. Human gene essentiality. Nat Rev Genet. 2018;19:51–62. - PubMed
-
- Battey J, Jordan E, Cox D, Dove W. An action plan for mouse genomics. Nat Genet. 1999;21:73–75. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

