Quantification of pulmonary involvement in COVID-19 pneumonia by means of a cascade of two U-nets: training and assessment on multiple datasets using different annotation criteria
- PMID: 34698988
- PMCID: PMC8547130
- DOI: 10.1007/s11548-021-02501-2
Quantification of pulmonary involvement in COVID-19 pneumonia by means of a cascade of two U-nets: training and assessment on multiple datasets using different annotation criteria
Abstract
Purpose: This study aims at exploiting artificial intelligence (AI) for the identification, segmentation and quantification of COVID-19 pulmonary lesions. The limited data availability and the annotation quality are relevant factors in training AI-methods. We investigated the effects of using multiple datasets, heterogeneously populated and annotated according to different criteria.
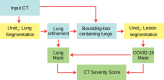
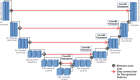
Methods: We developed an automated analysis pipeline, the LungQuant system, based on a cascade of two U-nets. The first one (U-net[Formula: see text]) is devoted to the identification of the lung parenchyma; the second one (U-net[Formula: see text]) acts on a bounding box enclosing the segmented lungs to identify the areas affected by COVID-19 lesions. Different public datasets were used to train the U-nets and to evaluate their segmentation performances, which have been quantified in terms of the Dice Similarity Coefficients. The accuracy in predicting the CT-Severity Score (CT-SS) of the LungQuant system has been also evaluated.
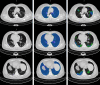
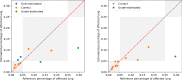
Results: Both the volumetric DSC (vDSC) and the accuracy showed a dependency on the annotation quality of the released data samples. On an independent dataset (COVID-19-CT-Seg), both the vDSC and the surface DSC (sDSC) were measured between the masks predicted by LungQuant system and the reference ones. The vDSC (sDSC) values of 0.95±0.01 and 0.66±0.13 (0.95±0.02 and 0.76±0.18, with 5 mm tolerance) were obtained for the segmentation of lungs and COVID-19 lesions, respectively. The system achieved an accuracy of 90% in CT-SS identification on this benchmark dataset.
Conclusion: We analysed the impact of using data samples with different annotation criteria in training an AI-based quantification system for pulmonary involvement in COVID-19 pneumonia. In terms of vDSC measures, the U-net segmentation strongly depends on the quality of the lesion annotations. Nevertheless, the CT-SS can be accurately predicted on independent test sets, demonstrating the satisfactory generalization ability of the LungQuant.
Keywords: COVID-19; Chest Computed Tomography; Ground-glass opacities; Machine Learning; Segmentation; U-net.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Quantification of pulmonary involvement in COVID-19 pneumonia: an upgrade of the LungQuant software for lung CT segmentation.Eur Phys J Plus. 2023;138(4):326. doi: 10.1140/epjp/s13360-023-03896-4. Epub 2023 Apr 11. Eur Phys J Plus. 2023. PMID: 37064789 Free PMC article.
-
A multicenter evaluation of a deep learning software (LungQuant) for lung parenchyma characterization in COVID-19 pneumonia.Eur Radiol Exp. 2023 Apr 10;7(1):18. doi: 10.1186/s41747-023-00334-z. Eur Radiol Exp. 2023. PMID: 37032383 Free PMC article.
-
Association of AI quantified COVID-19 chest CT and patient outcome.Int J Comput Assist Radiol Surg. 2021 Mar;16(3):435-445. doi: 10.1007/s11548-020-02299-5. Epub 2021 Jan 23. Int J Comput Assist Radiol Surg. 2021. PMID: 33484428 Free PMC article.
-
Human-in-the-Loop-A Deep Learning Strategy in Combination with a Patient-Specific Gaussian Mixture Model Leads to the Fast Characterization of Volumetric Ground-Glass Opacity and Consolidation in the Computed Tomography Scans of COVID-19 Patients.J Clin Med. 2024 Sep 4;13(17):5231. doi: 10.3390/jcm13175231. J Clin Med. 2024. PMID: 39274444 Free PMC article.
-
COVID-19 Imaging-based AI Research - A Literature Review.Curr Med Imaging. 2022;18(5):496-508. doi: 10.2174/1573405617666210902103729. Curr Med Imaging. 2022. PMID: 34473619 Review.
Cited by
-
COVLIAS 1.0Lesion vs. MedSeg: An Artificial Intelligence Framework for Automated Lesion Segmentation in COVID-19 Lung Computed Tomography Scans.Diagnostics (Basel). 2022 May 21;12(5):1283. doi: 10.3390/diagnostics12051283. Diagnostics (Basel). 2022. PMID: 35626438 Free PMC article.
-
The Evolution of Artificial Intelligence in Medical Imaging: From Computer Science to Machine and Deep Learning.Cancers (Basel). 2024 Nov 1;16(21):3702. doi: 10.3390/cancers16213702. Cancers (Basel). 2024. PMID: 39518140 Free PMC article. Review.
-
Understanding the Impact of Evaluation Metrics in Kinetic Models for Consensus-Based Segmentation.Entropy (Basel). 2025 Feb 1;27(2):149. doi: 10.3390/e27020149. Entropy (Basel). 2025. PMID: 40003146 Free PMC article.
-
CompositIA: an open-source automated quantification tool for body composition scores from thoraco-abdominal CT scans.Eur Radiol Exp. 2025 Jan 29;9(1):12. doi: 10.1186/s41747-025-00552-7. Eur Radiol Exp. 2025. PMID: 39881078 Free PMC article.
-
Self-paced Multi-view Learning for CT-based severity assessment of COVID-19.Biomed Signal Process Control. 2023 May;83:104672. doi: 10.1016/j.bspc.2023.104672. Epub 2023 Feb 8. Biomed Signal Process Control. 2023. PMID: 36777556 Free PMC article.
References
-
- An P, Xu S, Harmon SA, Turkbey EB, Sanford TH, Amalou A, Kassin M, Varble N, Blain M, Anderson V, Patella F, Carrafiello G, Turkbey BT, Wood BJ (2020) CT Images in COVID-19. 10.7937/tcia.2020.gqry-nc81
-
- Carotti M, Salaffi F, Sarzi-Puttini P, Agostini A, Borgheresi A, Minorati D, Galli M, Marotto D, Giovagnoni A. Chest CT features of coronavirus disease 2019 (COVID-19) pneumonia: key points for radiologists. Radiologia Medica. 2020;125(7):636–646. doi: 10.1007/s11547-020-01237-4. - DOI - PMC - PubMed
-
- Chollet F (2015) Keras. https://keras.io
-
- Fang X, Kruger U, Homayounieh F, Chao H, Zhang J, Digumarthy SR, Arru CD, Kalra MK, Yan P (2021) Association of AI quantified COVID-19 chest CT and patient outcome. International Journal of Computer Assisted Radiology and Surgery. 10.1007/s11548-020-02299-5. URL http://www.ncbi.nlm.nih.gov/pubmed/33484428 - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

