GATA4 induces liver fibrosis regression by deactivating hepatic stellate cells
- PMID: 34699385
- PMCID: PMC8675192
- DOI: 10.1172/jci.insight.150059
GATA4 induces liver fibrosis regression by deactivating hepatic stellate cells
Abstract
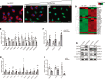
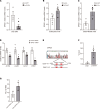
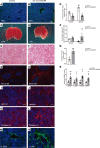
In response to liver injury, hepatic stellate cells activate and acquire proliferative and contractile features. The regression of liver fibrosis appears to involve the clearance of activated hepatic stellate cells, either by apoptosis or by reversion toward a quiescent-like state, a process called deactivation. Thus, deactivation of active hepatic stellate cells has emerged as a novel and promising therapeutic approach for liver fibrosis. However, our knowledge of the master regulators involved in the deactivation and/or activation of fibrotic hepatic stellate cells is still limited. The transcription factor GATA4 has been previously shown to play an important role in embryonic hepatic stellate cell quiescence. In this work, we show that lack of GATA4 in adult mice caused hepatic stellate cell activation and, consequently, liver fibrosis. During regression of liver fibrosis, Gata4 was reexpressed in deactivated hepatic stellate cells. Overexpression of Gata4 in hepatic stellate cells promoted liver fibrosis regression in CCl4-treated mice. GATA4 induced changes in the expression of fibrogenic and antifibrogenic genes, promoting hepatic stellate cell deactivation. Finally, we show that GATA4 directly repressed EPAS1 transcription in hepatic stellate cells and that stabilization of the HIF2α protein in hepatic stellate cells leads to liver fibrosis.
Keywords: Cell Biology; Fibrosis; Gastroenterology; Mouse models.
Figures



References
-
- Reeves HL, Friedman SL. Activation of hepatic stellate cells--a key issue in liver fibrosis. Front Biosci. 2002;7:d808–d826. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

