Single-cell sequencing unveils distinct immune microenvironments with CCR6-CCL20 crosstalk in human chronic pancreatitis
- PMID: 34702715
- PMCID: PMC9105403
- DOI: 10.1136/gutjnl-2021-324546
Single-cell sequencing unveils distinct immune microenvironments with CCR6-CCL20 crosstalk in human chronic pancreatitis
Abstract
Objective: Chronic pancreatitis (CP) is a potentially fatal disease of the exocrine pancreas, with no specific or effective approved therapies. Due to difficulty in accessing pancreas tissues, little is known about local immune responses or pathogenesis in human CP. We sought to characterise pancreatic immune responses using tissues derived from patients with different aetiologies of CP and non-CP organ donors in order to identify key signalling molecules associated with human CP.
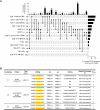
Design: We performed single-cell level cellular indexing of transcriptomes and epitopes by sequencing and T-cell receptor (TCR) sequencing of pancreatic immune cells isolated from organ donors, hereditary and idiopathic patients with CP who underwent total pancreatectomy. We validated gene expression data by performing flow cytometry and functional assays in a second patient with CP cohort.
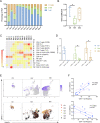
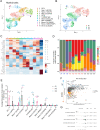
Results: Deep single-cell sequencing revealed distinct immune characteristics and significantly enriched CCR6+ CD4+ T cells in hereditary compared with idiopathic CP. In hereditary CP, a reduction in T-cell clonality was observed due to the increased CD4+ T (Th) cells that replaced tissue-resident CD8+ T cells. Shared TCR clonotype analysis among T-cell lineages also unveiled unique interactions between CCR6+ Th and Th1 subsets, and TCR clustering analysis showed unique common antigen binding motifs in hereditary CP. In addition, we observed a significant upregulation of the CCR6 ligand (CCL20) expression among monocytes in hereditary CP as compared with those in idiopathic CP. The functional significance of CCR6 expression in CD4+ T cells was confirmed by flow cytometry and chemotaxis assay.
Conclusion: Single-cell sequencing with pancreatic immune cells in human CP highlights pancreas-specific immune crosstalk through the CCR6-CCL20 axis, a signalling pathway that might be leveraged as a potential future target in human hereditary CP.
Keywords: RNA expression; T-cell receptor; chronic pancreatitis; immunology; pancreatitis.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
